Pyruvate kinase 2 from Synechocystis sp. PCC 6803 increased substrate affinity via glucose-6-phosphate and ribose-5-phosphate for phosphoenolpyruvate consumption
- PMID: 38758412
- PMCID: PMC11101554
- DOI: 10.1007/s11103-023-01401-0
Pyruvate kinase 2 from Synechocystis sp. PCC 6803 increased substrate affinity via glucose-6-phosphate and ribose-5-phosphate for phosphoenolpyruvate consumption
Abstract
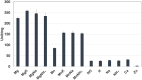
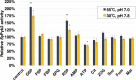
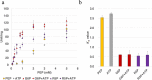
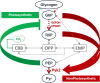
Pyruvate kinase (Pyk, EC 2.7.1.40) is a glycolytic enzyme that generates pyruvate and adenosine triphosphate (ATP) from phosphoenolpyruvate (PEP) and adenosine diphosphate (ADP), respectively. Pyk couples pyruvate and tricarboxylic acid metabolisms. Synechocystis sp. PCC 6803 possesses two pyk genes (encoded pyk1, sll0587 and pyk2, sll1275). A previous study suggested that pyk2 and not pyk1 is essential for cell viability; however, its biochemical analysis is yet to be performed. Herein, we biochemically analyzed Synechocystis Pyk2 (hereafter, SyPyk2). The optimum pH and temperature of SyPyk2 were 7.0 and 55 °C, respectively, and the Km values for PEP and ADP under optimal conditions were 1.5 and 0.053 mM, respectively. SyPyk2 is activated in the presence of glucose-6-phosphate (G6P) and ribose-5-phosphate (R5P); however, it remains unaltered in the presence of adenosine monophosphate (AMP) or fructose-1,6-bisphosphate. These results indicate that SyPyk2 is classified as PykA type rather than PykF, stimulated by sugar monophosphates, such as G6P and R5P, but not by AMP. SyPyk2, considering substrate affinity and effectors, can play pivotal roles in sugar catabolism under nonphotosynthetic conditions.
Keywords: Synechocystis sp. PCC 6803; Glycolysis; Microalgae; Pyruvate kinase.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
In Silico Study of the Structure and Ligand Preference of Pyruvate Kinases from Cyanobacterium Synechocystis sp. PCC 6803.Appl Biochem Biotechnol. 2021 Nov;193(11):3651-3671. doi: 10.1007/s12010-021-03630-9. Epub 2021 Aug 4. Appl Biochem Biotechnol. 2021. PMID: 34347252
-
Catabolite regulation analysis of Escherichia coli for acetate overflow mechanism and co-consumption of multiple sugars based on systems biology approach using computer simulation.J Biotechnol. 2013 Oct 20;168(2):155-73. doi: 10.1016/j.jbiotec.2013.06.023. Epub 2013 Jul 10. J Biotechnol. 2013. PMID: 23850830
-
Minimized Dark Consumption of Calvin Cycle Intermediates Facilitates the Initiation of Photosynthesis in Synechocystis sp. PCC 6803.Plant Cell Physiol. 2024 Dec 6;65(11):1812-1820. doi: 10.1093/pcp/pcae102. Plant Cell Physiol. 2024. PMID: 39238237
-
Central Role of Pyruvate Kinase in Carbon Co-catabolism of Mycobacterium tuberculosis.J Biol Chem. 2016 Mar 25;291(13):7060-9. doi: 10.1074/jbc.M115.707430. Epub 2016 Feb 8. J Biol Chem. 2016. PMID: 26858255 Free PMC article.
-
From genome to enzyme: analysis of key glycolytic and oxidative pentose-phosphate pathway enzymes in the cyanobacterium Synechocystis sp. PCC 6803.Plant Cell Physiol. 2003 Jul;44(7):758-63. doi: 10.1093/pcp/pcg086. Plant Cell Physiol. 2003. PMID: 12881504
Cited by
-
Development of a CRISPR activation system for targeted gene upregulation in Synechocystis sp. PCC 6803.Commun Biol. 2025 May 21;8(1):772. doi: 10.1038/s42003-025-08164-y. Commun Biol. 2025. PMID: 40399557 Free PMC article.
References
-
- Abernathy MH, Yu J, Ma F, Liberton M, Ungerer J, Hollinshead WD, Gopalakrishnan S, He L, Maranas CD, Pakrasi HB, Allen DK, Tang YJ. Deciphering cyanobacterial phenotypes for fast photoautotrophic growth via isotopically nonstationary metabolic flux analysis. Biotechnol Biofuels. 2017;10:273. doi: 10.1186/s13068-017-0958-y. - DOI - PMC - PubMed
-
- Allison W, Corey DB, Ashok P, Christie AMP (2019) A comprehensive time-course metabolite profiling of the model cyanobacterium Synechocystis sp. PCC 6803 under diurnal light:dark cycles - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

