DNA choreography: correlating mobility and organization of DNA across different resolutions from loops to chromosomes
- PMID: 38758428
- PMCID: PMC11227476
- DOI: 10.1007/s00418-024-02285-x
DNA choreography: correlating mobility and organization of DNA across different resolutions from loops to chromosomes
Abstract
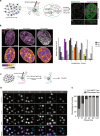
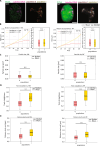
The dynamics of DNA in the cell nucleus plays a role in cellular processes and fates but the interplay of DNA mobility with the hierarchical levels of DNA organization is still underexplored. Here, we made use of DNA replication to directly label genomic DNA in an unbiased genome-wide manner. This was followed by live-cell time-lapse microscopy of the labeled DNA combining imaging at different resolutions levels simultaneously and allowing one to trace DNA motion across organization levels within the same cells. Quantification of the labeled DNA segments at different microscopic resolution levels revealed sizes comparable to the ones reported for DNA loops using 3D super-resolution microscopy, topologically associated domains (TAD) using 3D widefield microscopy, and also entire chromosomes. By employing advanced chromatin tracking and image registration, we discovered that DNA exhibited higher mobility at the individual loop level compared to the TAD level and even less at the chromosome level. Additionally, our findings indicate that chromatin movement, regardless of the resolution, slowed down during the S phase of the cell cycle compared to the G1/G2 phases. Furthermore, we found that a fraction of DNA loops and TADs exhibited directed movement with the majority depicting constrained movement. Our data also indicated spatial mobility differences with DNA loops and TADs at the nuclear periphery and the nuclear interior exhibiting lower velocity and radius of gyration than the intermediate locations. On the basis of these insights, we propose that there is a link between DNA mobility and its organizational structure including spatial distribution, which impacts cellular processes.
Keywords: Image registration; Live cell DNA labeling; Motion analysis; Single particle tracking; Super-resolution microscopy; Widefield microscopy.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Transcription-coupled structural dynamics of topologically associating domains regulate replication origin efficiency.Genome Biol. 2021 Jul 12;22(1):206. doi: 10.1186/s13059-021-02424-w. Genome Biol. 2021. PMID: 34253239 Free PMC article.
-
Super-resolution visualization and modeling of human chromosomal regions reveals cohesin-dependent loop structures.Genome Biol. 2021 May 11;22(1):150. doi: 10.1186/s13059-021-02343-w. Genome Biol. 2021. PMID: 33975635 Free PMC article.
-
Replisome loading reduces chromatin motion independent of DNA synthesis.Elife. 2023 Oct 31;12:RP87572. doi: 10.7554/eLife.87572. Elife. 2023. PMID: 37906089 Free PMC article.
-
Simulation of different three-dimensional polymer models of interphase chromosomes compared to experiments-an evaluation and review framework of the 3D genome organization.Semin Cell Dev Biol. 2019 Jun;90:19-42. doi: 10.1016/j.semcdb.2018.07.012. Epub 2018 Aug 24. Semin Cell Dev Biol. 2019. PMID: 30125668 Review.
-
Recent evidence that TADs and chromatin loops are dynamic structures.Nucleus. 2018 Jan 1;9(1):20-32. doi: 10.1080/19491034.2017.1389365. Epub 2017 Dec 14. Nucleus. 2018. PMID: 29077530 Free PMC article. Review.
Cited by
-
Seeing genomes.Histochem Cell Biol. 2024 Jul;162(1-2):1-2. doi: 10.1007/s00418-024-02301-0. Histochem Cell Biol. 2024. PMID: 38850309 No abstract available.
References
-
- Arthur D, Vassilvitskii S. k-means++: The advantages of careful seeding. In Soda. 2007;7:1027–1035.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

