Real-time forecasting of COVID-19-related hospital strain in France using a non-Markovian mechanistic model
- PMID: 38758962
- PMCID: PMC11139328
- DOI: 10.1371/journal.pcbi.1012124
Real-time forecasting of COVID-19-related hospital strain in France using a non-Markovian mechanistic model
Abstract
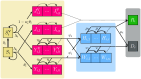
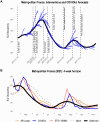
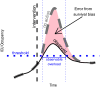
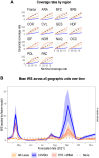
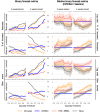
Projects such as the European Covid-19 Forecast Hub publish forecasts on the national level for new deaths, new cases, and hospital admissions, but not direct measurements of hospital strain like critical care bed occupancy at the sub-national level, which is of particular interest to health professionals for planning purposes. We present a sub-national French framework for forecasting hospital strain based on a non-Markovian compartmental model, its associated online visualisation tool and a retrospective evaluation of the real-time forecasts it provided from January to December 2021 by comparing to three baselines derived from standard statistical forecasting methods (a naive model, auto-regression, and an ensemble of exponential smoothing and ARIMA). In terms of median absolute error for forecasting critical care unit occupancy at the two-week horizon, our model only outperformed the naive baseline for 4 out of 14 geographical units and underperformed compared to the ensemble baseline for 5 of them at the 90% confidence level (n = 38). However, for the same level at the 4 week horizon, our model was never statistically outperformed for any unit despite outperforming the baselines 10 times spanning 7 out of 14 geographical units. This implies modest forecasting utility for longer horizons which may justify the application of non-Markovian compartmental models in the context of hospital-strain surveillance for future pandemics.
Copyright: © 2024 Massey et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Becker AD, Grantz KH, Hegde ST, Bérubé S, Cummings DAT, Wesolowski A. Development and dissemination of infectious disease dynamic transmission models during the COVID-19 pandemic: what can we learn from other pathogens and how can we move forward? Lancet Digit Health. 2021;3(1):e41–e50. doi: 10.1016/S2589-7500(20)30268-5 - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

