Connecting Species-Specific Extents of Genome Reduction in Mitochondria and Plastids
- PMID: 38758976
- PMCID: PMC11144018
- DOI: 10.1093/molbev/msae097
Connecting Species-Specific Extents of Genome Reduction in Mitochondria and Plastids
Abstract
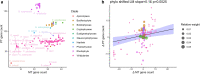
Mitochondria and plastids have both dramatically reduced their genomes since the endosymbiotic events that created them. The similarities and differences in the evolution of the two organelle genome types have been the target of discussion and investigation for decades. Ongoing work has suggested that similar mechanisms may modulate the reductive evolution of the two organelles in a given species, but quantitative data and statistical analyses exploring this picture remain limited outside of some specific cases like parasitism. Here, we use cross-eukaryote organelle genome data to explore evidence for coevolution of mitochondrial and plastid genome reduction. Controlling for differences between clades and pseudoreplication due to relatedness, we find that extents of mtDNA and ptDNA gene retention are related to each other across taxa, in a generally positive correlation that appears to differ quantitatively across eukaryotes, for example, between algal and nonalgal species. We find limited evidence for coevolution of specific mtDNA and ptDNA gene pairs, suggesting that the similarities between the two organelle types may be due mainly to independent responses to consistent evolutionary drivers.
Keywords: chloroplasts; genome evolution; mitochondria; organelle DNA; phylogenetic comparison.
© The Author(s) 2024. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures


References
-
- Ahyong S, Boyko CB, Bailly N, Bernot J, Bieler R, Brandão SN, Daly M, De Grave S, Gofas S, Hernandez F, et al. 2023. World Register of Marine Species (WoRMS). WoRMS Editorial Board. [accessed 2023 Sep]. https://www.marinespecies.org.
-
- Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015:67(1):1–48. 10.18637/jss.v067.i01. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

