PQSDC: a parallel lossless compressor for quality scores data via sequences partition and run-length prediction mapping
- PMID: 38759114
- PMCID: PMC11139522
- DOI: 10.1093/bioinformatics/btae323
PQSDC: a parallel lossless compressor for quality scores data via sequences partition and run-length prediction mapping
Abstract
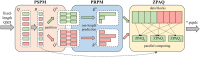
Motivation: The quality scores data (QSD) account for 70% in compressed FastQ files obtained from the short and long reads sequencing technologies. Designing effective compressors for QSD that counterbalance compression ratio, time cost, and memory consumption is essential in scenarios such as large-scale genomics data sharing and long-term data backup. This study presents a novel parallel lossless QSD-dedicated compression algorithm named PQSDC, which fulfills the above requirements well. PQSDC is based on two core components: a parallel sequences-partition model designed to reduce peak memory consumption and time cost during compression and decompression processes, as well as a parallel four-level run-length prediction mapping model to enhance compression ratio. Besides, the PQSDC algorithm is also designed to be highly concurrent using multicore CPU clusters.
Results: We evaluate PQSDC and four state-of-the-art compression algorithms on 27 real-world datasets, including 61.857 billion QSD characters and 632.908 million QSD sequences. (1) For short reads, compared to baselines, the maximum improvement of PQSDC reaches 7.06% in average compression ratio, and 8.01% in weighted average compression ratio. During compression and decompression, the maximum total time savings of PQSDC are 79.96% and 84.56%, respectively; the maximum average memory savings are 68.34% and 77.63%, respectively. (2) For long reads, the maximum improvement of PQSDC reaches 12.51% and 13.42% in average and weighted average compression ratio, respectively. The maximum total time savings during compression and decompression are 53.51% and 72.53%, respectively; the maximum average memory savings are 19.44% and 17.42%, respectively. (3) Furthermore, PQSDC ranks second in compression robustness among the tested algorithms, indicating that it is less affected by the probability distribution of the QSD collections. Overall, our work provides a promising solution for QSD parallel compression, which balances storage cost, time consumption, and memory occupation primely.
Availability and implementation: The proposed PQSDC compressor can be downloaded from https://github.com/fahaihi/PQSDC.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures
Similar articles
-
PMFFRC: a large-scale genomic short reads compression optimizer via memory modeling and redundant clustering.BMC Bioinformatics. 2023 Nov 30;24(1):454. doi: 10.1186/s12859-023-05566-9. BMC Bioinformatics. 2023. PMID: 38036969 Free PMC article.
-
SPRING: a next-generation compressor for FASTQ data.Bioinformatics. 2019 Aug 1;35(15):2674-2676. doi: 10.1093/bioinformatics/bty1015. Bioinformatics. 2019. PMID: 30535063 Free PMC article.
-
LCQS: an efficient lossless compression tool of quality scores with random access functionality.BMC Bioinformatics. 2020 Mar 18;21(1):109. doi: 10.1186/s12859-020-3428-7. BMC Bioinformatics. 2020. PMID: 32183707 Free PMC article.
-
Vertical lossless genomic data compression tools for assembled genomes: A systematic literature review.PLoS One. 2020 May 26;15(5):e0232942. doi: 10.1371/journal.pone.0232942. eCollection 2020. PLoS One. 2020. PMID: 32453750 Free PMC article.
-
Multi-file dynamic compression method based on classification algorithm in DNA storage.Med Biol Eng Comput. 2024 Dec;62(12):3623-3635. doi: 10.1007/s11517-024-03156-2. Epub 2024 Jun 26. Med Biol Eng Comput. 2024. PMID: 38922373 Review.
References
-
- Cánovas R, Moffat A, Turpin A. et al. Lossy compression of quality scores in genomic data. Bioinformatics 2014;30:2130–6. - PubMed