The F-actin bundler SWAP-70 promotes tumor metastasis
- PMID: 38760173
- PMCID: PMC11101836
- DOI: 10.26508/lsa.202302307
The F-actin bundler SWAP-70 promotes tumor metastasis
Abstract
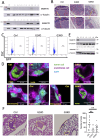
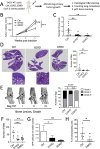
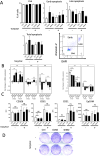
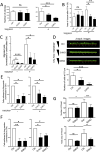
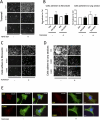
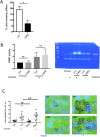
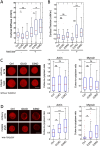
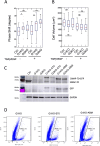
Dynamic rearrangements of the F-actin cytoskeleton are a hallmark of tumor metastasis. Thus, proteins that govern F-actin rearrangements are of major interest for understanding metastasis and potential therapies. We hypothesized that the unique F-actin binding and bundling protein SWAP-70 contributes importantly to metastasis. Orthotopic, ectopic, and short-term tail vein injection mouse breast and lung cancer models revealed a strong positive dependence of lung and bone metastasis on SWAP-70. Breast cancer cell growth, migration, adhesion, and invasion assays revealed SWAP-70's key role in these metastasis-related cell features and the requirement for SWAP-70 to bind F-actin. Biophysical experiments showed that tumor cell stiffness and deformability are negatively modulated by SWAP-70. Together, we present a hitherto undescribed, unique F-actin modulator as an important contributor to tumor metastasis.
© 2024 Chang et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


Similar articles
-
The switch-associated protein 70 (SWAP-70) bundles actin filaments and contributes to the regulation of F-actin dynamics.J Biol Chem. 2013 Oct 4;288(40):28687-703. doi: 10.1074/jbc.M113.461277. Epub 2013 Aug 6. J Biol Chem. 2013. PMID: 23921380 Free PMC article.
-
Mechanism of control of F-actin cortex architecture by SWAP-70.J Cell Sci. 2020 Jan 23;133(2):jcs233064. doi: 10.1242/jcs.233064. J Cell Sci. 2020. PMID: 31932501
-
Regulation of bone mass and osteoclast function depend on the F-actin modulator SWAP-70.J Bone Miner Res. 2012 Oct;27(10):2085-96. doi: 10.1002/jbmr.1670. J Bone Miner Res. 2012. PMID: 22648978
-
Juxtanuclear Drebrin-Enriched Zone.Adv Exp Med Biol. 2017;1006:329-336. doi: 10.1007/978-4-431-56550-5_19. Adv Exp Med Biol. 2017. PMID: 28865029 Review.
-
How does fascin promote cancer metastasis?FEBS J. 2021 Mar;288(5):1434-1446. doi: 10.1111/febs.15484. Epub 2020 Jul 23. FEBS J. 2021. PMID: 32657526 Free PMC article. Review.
References
-
- Bemmerlein L, Deniz IA, Karbanova J, Jacobi A, Drukewitz S, Link T, Gobel A, Sevenich L, Taubenberger AV, Wimberger P, et al. (2022) Decoding single cell morphology in osteotropic breast cancer cells for dissecting their migratory, molecular and biophysical heterogeneity. Cancers 14: 603. 10.3390/cancers14030603 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
