Mapping recurrent mosaic copy number variation in human neurons
- PMID: 38760338
- PMCID: PMC11101435
- DOI: 10.1038/s41467-024-48392-0
Mapping recurrent mosaic copy number variation in human neurons
Abstract
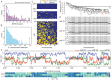
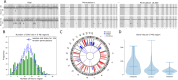
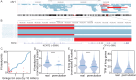
When somatic cells acquire complex karyotypes, they often are removed by the immune system. Mutant somatic cells that evade immune surveillance can lead to cancer. Neurons with complex karyotypes arise during neurotypical brain development, but neurons are almost never the origin of brain cancers. Instead, somatic mutations in neurons can bring about neurodevelopmental disorders, and contribute to the polygenic landscape of neuropsychiatric and neurodegenerative disease. A subset of human neurons harbors idiosyncratic copy number variants (CNVs, "CNV neurons"), but previous analyses of CNV neurons are limited by relatively small sample sizes. Here, we develop an allele-based validation approach, SCOVAL, to corroborate or reject read-depth based CNV calls in single human neurons. We apply this approach to 2,125 frontal cortical neurons from a neurotypical human brain. SCOVAL identifies 226 CNV neurons, which include a subclass of 65 CNV neurons with highly aberrant karyotypes containing whole or substantial losses on multiple chromosomes. Moreover, we find that CNV location appears to be nonrandom. Recurrent regions of neuronal genome rearrangement contain fewer, but longer, genes.
© 2024. The Author(s).
Conflict of interest statement
J.V.M. is an inventor on patent US6150160, is a paid consultant for Gilead Sciences, serves on the scientific advisory board of Tessera Therapeutics Inc. (where he is paid as a consultant, and has equity options), has licensed reagents to Merck Pharmaceutical, and recently served on the American Society of Human Genetics Board of Directors. The remaining authors declare no competing interests.
Figures

References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

