DDX5 inhibits hyaline cartilage fibrosis and degradation in osteoarthritis via alternative splicing and G-quadruplex unwinding
- PMID: 38760576
- PMCID: PMC11108786
- DOI: 10.1038/s43587-024-00624-0
DDX5 inhibits hyaline cartilage fibrosis and degradation in osteoarthritis via alternative splicing and G-quadruplex unwinding
Abstract
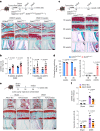
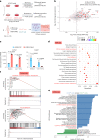
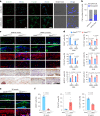
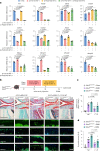
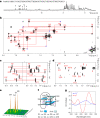
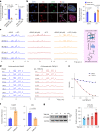
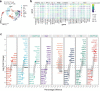
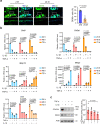
Hyaline cartilage fibrosis is typically considered an end-stage pathology of osteoarthritis (OA), which results in changes to the extracellular matrix. However, the mechanism behind this is largely unclear. Here, we found that the RNA helicase DDX5 was dramatically downregulated during the progression of OA. DDX5 deficiency increased fibrosis phenotype by upregulating COL1 expression and downregulating COL2 expression. In addition, loss of DDX5 aggravated cartilage degradation by inducing the production of cartilage-degrading enzymes. Chondrocyte-specific deletion of Ddx5 led to more severe cartilage lesions in the mouse OA model. Mechanistically, weakened DDX5 resulted in abundance of the Fn1-AS-WT and Plod2-AS-WT transcripts, which promoted expression of fibrosis-related genes (Col1, Acta2) and extracellular matrix degradation genes (Mmp13, Nos2 and so on), respectively. Additionally, loss of DDX5 prevented the unfolding Col2 promoter G-quadruplex, thereby reducing COL2 production. Together, our data suggest that strategies aimed at the upregulation of DDX5 hold significant potential for the treatment of cartilage fibrosis and degradation in OA.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures










References
MeSH terms
Substances
Grants and funding
- 91853109/National Natural Science Foundation of China (National Science Foundation of China)
- 81872877/National Natural Science Foundation of China (National Science Foundation of China)
- 81673436/National Natural Science Foundation of China (National Science Foundation of China)
- 32200995/National Natural Science Foundation of China (National Science Foundation of China)
- 21937005/National Natural Science Foundation of China (National Science Foundation of China)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

