Comparative 3D genome analysis between neural retina and retinal pigment epithelium reveals differential cis-regulatory interactions at retinal disease loci
- PMID: 38760655
- PMCID: PMC11100165
- DOI: 10.1186/s13059-024-03250-6
Comparative 3D genome analysis between neural retina and retinal pigment epithelium reveals differential cis-regulatory interactions at retinal disease loci
Abstract
Background: Vision depends on the interplay between photoreceptor cells of the neural retina and the underlying retinal pigment epithelium (RPE). Most genes involved in inherited retinal diseases display specific spatiotemporal expression within these interconnected retinal components through the local recruitment of cis-regulatory elements (CREs) in 3D nuclear space.
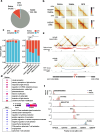
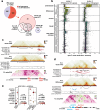
Results: To understand the role of differential chromatin architecture in establishing tissue-specific expression at inherited retinal disease loci, we mapped genome-wide chromatin interactions using in situ Hi-C and H3K4me3 HiChIP on neural retina and RPE/choroid from human adult donor eyes. We observed chromatin looping between active promoters and 32,425 and 8060 candidate CREs in the neural retina and RPE/choroid, respectively. A comparative 3D genome analysis between these two retinal tissues revealed that 56% of 290 known inherited retinal disease genes were marked by differential chromatin interactions. One of these was ABCA4, which is implicated in the most common autosomal recessive inherited retinal disease. We zoomed in on retina- and RPE-specific cis-regulatory interactions at the ABCA4 locus using high-resolution UMI-4C. Integration with bulk and single-cell epigenomic datasets and in vivo enhancer assays in zebrafish revealed tissue-specific CREs interacting with ABCA4.
Conclusions: Through comparative 3D genome mapping, based on genome-wide, promoter-centric, and locus-specific assays of human neural retina and RPE, we have shown that gene regulation at key inherited retinal disease loci is likely mediated by tissue-specific chromatin interactions. These findings do not only provide insight into tissue-specific regulatory landscapes at retinal disease loci, but also delineate the search space for non-coding genomic variation underlying unsolved inherited retinal diseases.
Keywords: ABCA4; Cis-regulatory element (CRE); 3D genome structure; Enhancer assay; Hi-C; HiChIP; Inherited retinal disease (IRD); Neural retina; Retinal pigment epithelium (RPE); UMI-4C.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
- BOF20/GOA/023/Ghent University Special Research Fund
- 813490/H2020 Marie Skłodowska-Curie Actions
- 813490/H2020 Marie Skłodowska-Curie Actions
- 813490/H2020 Marie Skłodowska-Curie Actions
- 813490/H2020 Marie Skłodowska-Curie Actions
- 813490/H2020 Marie Skłodowska-Curie Actions
- EJPRD19-234/European Joint Programme on Rare Diseases (EJP RD)
- EJPRD19-234/European Joint Programme on Rare Diseases (EJP RD)
- EJPRD19-234/European Joint Programme on Rare Diseases (EJP RD)
- EJPRD19-234/European Joint Programme on Rare Diseases (EJP RD)
- G0A9718N/Research Foundation Flanders (FWO)
- 1802220N/Research Foundation Flanders (FWO)
- 12D8523N/Research Foundation Flanders (FWO)
- DOC_00397/Junta de Andalucía
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

