Giant viral signatures on the Greenland ice sheet
- PMID: 38760842
- PMCID: PMC11100222
- DOI: 10.1186/s40168-024-01796-y
Giant viral signatures on the Greenland ice sheet
Abstract
Background: Dark pigmented snow and glacier ice algae on glaciers and ice sheets contribute to accelerating melt. The biological controls on these algae, particularly the role of viruses, remain poorly understood. Giant viruses, classified under the nucleocytoplasmic large DNA viruses (NCLDV) supergroup (phylum Nucleocytoviricota), are diverse and globally distributed. NCLDVs are known to infect eukaryotic cells in marine and freshwater environments, providing a biological control on the algal population in these ecosystems. However, there is very limited information on the diversity and ecosystem function of NCLDVs in terrestrial icy habitats.
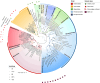
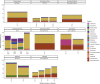
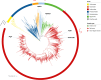
Results: In this study, we investigate for the first time giant viruses and their host connections on ice and snow habitats, such as cryoconite, dark ice, ice core, red and green snow, and genomic assemblies of five cultivated Chlorophyta snow algae. Giant virus marker genes were present in almost all samples; the highest abundances were recovered from red snow and the snow algae genomic assemblies, followed by green snow and dark ice. The variety of active algae and protists in these GrIS habitats containing NCLDV marker genes suggests that infection can occur on a range of eukaryotic hosts. Metagenomic data from red and green snow contained evidence of giant virus metagenome-assembled genomes from the orders Imitervirales, Asfuvirales, and Algavirales.
Conclusion: Our study highlights NCLDV family signatures in snow and ice samples from the Greenland ice sheet. Giant virus metagenome-assembled genomes (GVMAGs) were found in red snow samples, and related NCLDV marker genes were identified for the first time in snow algal culture genomic assemblies; implying a relationship between the NCLDVs and snow algae. Metatranscriptomic viral genes also aligned with metagenomic sequences, suggesting that NCLDVs are an active component of the microbial community and are potential "top-down" controls of the eukaryotic algal and protistan members. This study reveals the unprecedented presence of a diverse community of NCLDVs in a variety of glacial habitats dominated by algae.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Tanaka S, Takeuchi N, Miyairi M, Fujisawa Y, Kadota T, Shirakawa T, et al. Snow algal communities on glaciers in the Suntar-Khayata Mountain Range in eastern Siberia, Russia. Polar Sci. 2016;10:227–38. Available from: 10.1016/j.polar.2016.03.004.
MeSH terms
LinkOut - more resources
Full Text Sources

