Coaggregated E. faecalis with F. nucleatum regulated environmental stress responses and inflammatory effects
- PMID: 38761182
- PMCID: PMC11102388
- DOI: 10.1007/s00253-024-13172-9
Coaggregated E. faecalis with F. nucleatum regulated environmental stress responses and inflammatory effects
Abstract
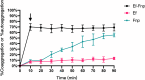
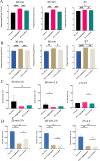
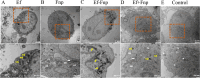
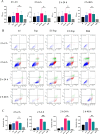
To investigate the cell-cell interactions of intergeneric bacterial species, the study detected the survival of Enterococcus faecalis (Ef) under monospecies or coaggregation state with Fusobacterium nucleatum subsp. polymorphum (Fnp) in environmental stress. Ef and Fnp infected the human macrophages with different forms (Ef and Fnp monospecies, Ef-Fnp coaggregates, Ef + Fnp cocultures) for exploring the immunoregulatory effects and the relevant molecular mechanisms. Meanwhile, the transcriptomic profiles of coaggregated Ef and Fnp were analyzed. Ef was shown to coaggregate with Fnp strongly in CAB within 90 min by forming multiplexes clumps. Coaggregation with Fnp reinforced Ef resistance against unfavorable conditions including alkaline, hypertonic, nutrient-starvation, and antibiotic challenges. Compared with monospecies and coculture species, the coaggregation of Ef and Fnp significantly facilitates both species to invade dTHP-1 cells and aid Ef to survive within the cells. Compared with coculture species, dual-species interaction of Ef and Fnp significantly decreased the levels of pro-inflammatory cytokines IL-6, TNF-α, and chemokines MCP-1 secreted by dTHP-1 cells and lessened the phosphorylation of p38, JNK, and p65 signaling pathways. The transcriptome sequencing results showed that 111 genes were differentially expressed or Ef-Fnp coaggregated species compared to Ef monospecies; 651 genes were differentially expressed for Fnp when coaggregation with Ef. The analysis of KEGG pathway showed that Ef differentially expressed genes (DEGs) were enriched in quorum sensing and arginine biosynthesis pathway; Fnp DEGs were differentially concentrated in lipopolysaccharide (LPS) biosynthesis, biofilm formation, and lysine degradation pathway compared to monospecies. KEY POINTS: • Coaggregated with Fnp aids Ef's survival in environmental stress, especially in root canals after endodontic treatment. • The coaggregation of Ef and Fnp may weaken the pro-inflammatory response and facilitate Ef to evade killed by macrophages. • The coaggregation between Ef and Fnp altered interspecies transcriptional profiles.
Keywords: Enterococcus faecalis; Fusobacterium nucleatum; Coaggregation; Host-pathogen interaction; Immunoregulatory effects; Transcriptomics.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no conflicting interests.
Figures






Similar articles
-
Interactions Between Streptococcus gordonii and Fusobacterium nucleatum Altered Bacterial Transcriptional Profiling and Attenuated the Immune Responses of Macrophages.Front Cell Infect Microbiol. 2022 Jan 7;11:783323. doi: 10.3389/fcimb.2021.783323. eCollection 2021. Front Cell Infect Microbiol. 2022. PMID: 35071038 Free PMC article.
-
Coaggregation interactions between oral and endodontic Enterococcus faecalis and bacterial species isolated from persistent apical periodontitis.J Endod. 2006 Oct;32(10):946-50. doi: 10.1016/j.joen.2006.03.023. Epub 2006 Jul 12. J Endod. 2006. PMID: 16982270
-
The Regulatory Effect of Coaggregation Between Fusobacterium nucleatum and Streptococcus gordonii on the Synergistic Virulence to Human Gingival Epithelial Cells.Front Cell Infect Microbiol. 2022 Apr 29;12:879423. doi: 10.3389/fcimb.2022.879423. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35573793 Free PMC article.
-
Aggregatibacter actinomycetemcomitans serotype f O-polysaccharide mediates coaggregation with Fusobacterium nucleatum.Oral Microbiol Immunol. 2008 Apr;23(2):127-30. doi: 10.1111/j.1399-302X.2007.00399.x. Oral Microbiol Immunol. 2008. PMID: 18279180
-
Transcriptional responses of Streptococcus gordonii and Fusobacterium nucleatum to coaggregation.Mol Oral Microbiol. 2018 Dec;33(6):450-464. doi: 10.1111/omi.12248. Mol Oral Microbiol. 2018. PMID: 30329223
Cited by
-
Interaction Between Enterococcus faecalis and Fusobacterium nucleatum Regulated Macrophage Transcriptional Profiling and Reprogrammed Cellular Immune and Metabolic Response.Microorganisms. 2025 Jun 11;13(6):1351. doi: 10.3390/microorganisms13061351. Microorganisms. 2025. PMID: 40572241 Free PMC article.
References
-
- Akcay M, Arslan H, Durmus N, Mese M, Capar ID (2016) Dentinal tubule penetration of AH plus, iRoot SP, MTA fillapex, and guttaflow bioseal root canal sealers after different final irrigation procedures: a confocal microscopic study. Lasers Surg Med 48(1):70–76. 10.1002/lsm.22446 - PubMed
-
- Baldassarri L, Bertuccini L, Creti R, Filippini P, Ammendolia MG, Koch S, Huebner J, Orefici G (2005) Glycosaminoglycans mediate invasion and survival of Enterococcus faecalis into macrophages. J Infect Dis 191(8):1253–1262. 10.1086/428778 - PubMed
-
- Cassenego AP, de Oliveira NE, Laport MS, Abranches J, Lemos JA, Giambiagi-deMarval M (2016) The CtsR regulator controls the expression of clpC, clpE and clpP and is required for the virulence of Enterococcus faecalis in an invertebrate model. Antonie Van Leeuwenhoek 109(9):1253–1259. 10.1007/s10482-016-0727-0 - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

