Genome-wide investigation of lncRNAs revealed their tight association with gastric cancer
- PMID: 38761291
- PMCID: PMC11102383
- DOI: 10.1007/s00432-024-05790-7
Genome-wide investigation of lncRNAs revealed their tight association with gastric cancer
Abstract
Background: Gastric cancer (GC) is a significant health issue globally, ranking as the fifth most common cancer with over 10,000 new cases reported annually. Long non-coding RNA (lncRNA) has emerged as a critical player in cellular functions, influencing GC's development, growth, metastasis, and prognosis. However, our understanding of lncRNA's role in the pathogenesis of GC remains limited. Therefore, it is particularly important to explore the relationship between lncRNA and gastric cancer.
Methods: we conducted a comprehensive analysis of RNA sequencing data from the GEO database and stomach adenocarcinoma (STAD) data from the TCGA database to identify lncRNAs that exhibit altered expression levels in GC and the mechanisms underlying lncRNA-mediated transcription and post-transcriptional regulation were explored.
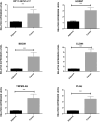
Results: This study uncovered 94 lncRNAs with differential expression and, through co-expression analysis, linked these to 1508 differentially expressed genes (DEGs). GO functional enrichment analysis highlighted that these DEGs are involved in critical pathways, such as cell adhesion and the positive regulation of cell migration. By establishing a lncRNA-miRNA-mRNA regulatory network, we found that the ceRNA mechanism, particularly involving RP11-357H14.17 and CTD-2377D24.4, could play a role in GC progression. Experimental validation of selected differentially expressed lncRNAs and mRNAs (including RP11-357H14.17-CLDN1, BBOX1, TRPM2-AS, CLDN1, PLAU, HOXB7) confirmed the RNA-seq results.
Conclusions: Overall, our findings highlight the critical role of the lncRNA-mRNA regulatory network in the development and progression of GC, offering potential biomarkers for diagnosis and targets for innovative treatment strategies.
Keywords: Co-expression network; Gastric cancer; lncRNA; mRNA.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




References
-
- Alsina M, Arrazubi V, Diez M, Tabernero J (2023) Current developments in gastric cancer: from molecular profiling to treatment strategy. Nat Rev Gastroenterol Hepatol 20(3):155–170. 10.1038/s41575-022-00703-w. (Epub 2022 Nov 7 PMID: 36344677) - PubMed
-
- Amirmahani F, Asadi MH, Jannat AF (2023b) LncRNA MIAT promotes the proliferation and invasion of colorectal cancer via suppressing apoptosis and senescence. Middle East J Cancer 14(2):219–229. 10.30476/mejc.2022.92233.1651
-
- Amirmahani F, Vallian S, Asadi MH (2023a) The LncRNA MIAT is identified as a regulator of stemness-associated transcript in glioma. Mol Biol Rep 50(1):517–530. 10.1007/s11033-022-07962-5. (Epub 2022 Nov 9 PMID: 36352177) - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

