Compensation between FOXP transcription factors maintains proper striatal function
- PMID: 38761373
- PMCID: PMC11234887
- DOI: 10.1016/j.celrep.2024.114257
Compensation between FOXP transcription factors maintains proper striatal function
Abstract
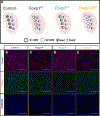
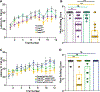
Spiny projection neurons (SPNs) of the striatum are critical in integrating neurochemical information to coordinate motor and reward-based behavior. Mutations in the regulatory transcription factors expressed in SPNs can result in neurodevelopmental disorders (NDDs). Paralogous transcription factors Foxp1 and Foxp2, which are both expressed in the dopamine receptor 1 (D1) expressing SPNs, are known to have variants implicated in NDDs. Utilizing mice with a D1-SPN-specific loss of Foxp1, Foxp2, or both and a combination of behavior, electrophysiology, and cell-type-specific genomic analysis, loss of both genes results in impaired motor and social behavior as well as increased firing of the D1-SPNs. Differential gene expression analysis implicates genes involved in autism risk, electrophysiological properties, and neuronal development and function. Viral-mediated re-expression of Foxp1 into the double knockouts is sufficient to restore electrophysiological and behavioral deficits. These data indicate complementary roles between Foxp1 and Foxp2 in the D1-SPNs.
Keywords: CP: Molecular biology; CP: Neuroscience; FOXP1; FOXP2; autism; neurogenomics; spiny projection neurons; striatum.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





Update of
-
Compensation between FOXP transcription factors maintains proper striatal function.bioRxiv [Preprint]. 2023 Jun 26:2023.06.26.546567. doi: 10.1101/2023.06.26.546567. bioRxiv. 2023. Update in: Cell Rep. 2024 May 28;43(5):114257. doi: 10.1016/j.celrep.2024.114257. PMID: 37425820 Free PMC article. Updated. Preprint.
References
-
- Gerfen CR (1992). The neostriatal mosaic: multiple levels of compartmental organization. Trends Neurosci. 15, 133–139. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

