Genome-scale analysis of interactions between genetic perturbations and natural variation
- PMID: 38762544
- PMCID: PMC11102447
- DOI: 10.1038/s41467-024-48626-1
Genome-scale analysis of interactions between genetic perturbations and natural variation
Abstract
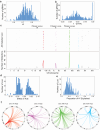
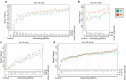
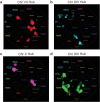
Interactions between genetic perturbations and segregating loci can cause perturbations to show different phenotypic effects across genetically distinct individuals. To study these interactions on a genome scale in many individuals, we used combinatorial DNA barcode sequencing to measure the fitness effects of 8046 CRISPRi perturbations targeting 1721 distinct genes in 169 yeast cross progeny (or segregants). We identified 460 genes whose perturbation has different effects across segregants. Several factors caused perturbations to show variable effects, including baseline segregant fitness, the mean effect of a perturbation across segregants, and interacting loci. We mapped 234 interacting loci and found four hub loci that interact with many different perturbations. Perturbations that interact with a given hub exhibit similar epistatic relationships with the hub and show enrichment for cellular processes that may mediate these interactions. These results suggest that an individual's response to perturbations is shaped by a network of perturbation-locus interactions that cannot be measured by approaches that examine perturbations or natural variation alone.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Update of
-
Genome-scale analysis of interactions between genetic perturbations and natural variation.bioRxiv [Preprint]. 2024 Jan 16:2023.05.06.539663. doi: 10.1101/2023.05.06.539663. bioRxiv. 2024. Update in: Nat Commun. 2024 May 18;15(1):4234. doi: 10.1038/s41467-024-48626-1. PMID: 38293072 Free PMC article. Updated. Preprint.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

