NAT10-mediated upregulation of GAS5 facilitates immune cell infiltration in non-small cell lung cancer via the MYBBP1A-p53/IRF1/type I interferon signaling axis
- PMID: 38762546
- PMCID: PMC11102450
- DOI: 10.1038/s41420-024-01997-2
NAT10-mediated upregulation of GAS5 facilitates immune cell infiltration in non-small cell lung cancer via the MYBBP1A-p53/IRF1/type I interferon signaling axis
Abstract
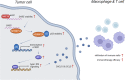
Interactions of tumor cells with immune cells in the tumor microenvironment play an important role during malignancy progression. We previously identified that GAS5 inhibited tumor development by suppressing proliferation of tumor cells in non-small cell lung cancer (NSCLC). Herein, we discovered a tumor-suppressing role for tumor cell-derived GAS5 in regulating tumor microenvironment. GAS5 positively coordinated with the infiltration of macrophages and T cells in NSCLC clinically, and overexpression of GAS5 promoted macrophages and T cells recruitment both in vitro and in vivo. Mechanistically, GAS5 stabilized p53 by directly binding to MYBBP1A and facilitating MYBBP1A-p53 interaction, and enhanced p53-mediated transcription of IRF1, which activated type I interferon signaling and increased the production of downstream CXCL10 and CCL5. We also found that activation of type I interferon signaling was associated with better immunotherapy efficacy in NSCLC. Furthermore, the stability of GAS5 was regulated by NAT10, the key enzyme responsible for N4-acetylcytidine (ac4C) modification, which bound to GAS5 and mediated its ac4C modification. Collectively, tumor cell-derived GAS5 could activate type I interferon signaling via the MYBBP1A-p53/IRF1 axis, promoting immune cell infiltration and potentially correlating with immunotherapy efficacy, which suppressed NSCLC progression. Our results suggested GAS5 as a promising predictive marker and potential therapeutic target for combination therapy in NSCLC. A schematic diagram demonstrating the regulatory effect of GAS5 on immune cell infiltration by activating type I interferon signaling via MYBBP1A-p53/IRF1 axis in non-small cell lung cancer. IFN, interferon.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






Similar articles
-
Helicobacter pylori-induced NAT10 stabilizes MDM2 mRNA via RNA acetylation to facilitate gastric cancer progression.J Exp Clin Cancer Res. 2023 Jan 6;42(1):9. doi: 10.1186/s13046-022-02586-w. J Exp Clin Cancer Res. 2023. PMID: 36609449 Free PMC article.
-
NAT10/ac4C/JunB facilitates TNBC malignant progression and immunosuppression by driving glycolysis addiction.J Exp Clin Cancer Res. 2024 Oct 4;43(1):278. doi: 10.1186/s13046-024-03200-x. J Exp Clin Cancer Res. 2024. PMID: 39363363 Free PMC article.
-
The positive feedback loop of the NAT10/Mybbp1a/p53 axis promotes cardiomyocyte ferroptosis to exacerbate cardiac I/R injury.Redox Biol. 2024 Jun;72:103145. doi: 10.1016/j.redox.2024.103145. Epub 2024 Apr 2. Redox Biol. 2024. PMID: 38583415 Free PMC article.
-
Ailanthone inhibits non-small cell lung cancer growth and metastasis through targeting UPF1/GAS5/ULK1 signaling pathway.Phytomedicine. 2024 Jun;128:155333. doi: 10.1016/j.phymed.2023.155333. Epub 2023 Dec 31. Phytomedicine. 2024. PMID: 38518633
-
Mechanisms of NAT10 as ac4C writer in diseases.Mol Ther Nucleic Acids. 2023 Apr 3;32:359-368. doi: 10.1016/j.omtn.2023.03.023. eCollection 2023 Jun 13. Mol Ther Nucleic Acids. 2023. PMID: 37128278 Free PMC article. Review.
Cited by
-
Investigating the role of exosomal long non-coding RNAs in drug resistance within female reproductive system cancers.Front Cell Dev Biol. 2025 Jan 24;13:1485422. doi: 10.3389/fcell.2025.1485422. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 39925739 Free PMC article. Review.
-
NAT10 Knockdown Improves Cisplatin Sensitivity in Non-Small Cell Lung Cancer by Inhibiting the TRIM44/PI3K/AKT Pathway.Thorac Cancer. 2025 May;16(9):e70079. doi: 10.1111/1759-7714.70079. Thorac Cancer. 2025. PMID: 40324967 Free PMC article.
-
RNA N4-acetylcytidine modification and its role in health and diseases.MedComm (2020). 2025 Jan 3;6(1):e70015. doi: 10.1002/mco2.70015. eCollection 2025 Jan. MedComm (2020). 2025. PMID: 39764566 Free PMC article. Review.
-
NAT10 promotes radiotherapy resistance in non-small cell lung cancer by regulating KPNB1-mediated PD-L1 nuclear translocation.Open Life Sci. 2025 Mar 18;20(1):20251065. doi: 10.1515/biol-2025-1065. eCollection 2025. Open Life Sci. 2025. PMID: 40109769 Free PMC article.
-
The Multifaceted Role of lncRNA GAS5: A Pan-Cancer Analysis of Its Diagnostic, Prognostic, and Therapeutic Potential.Cureus. 2025 Jan 16;17(1):e77527. doi: 10.7759/cureus.77527. eCollection 2025 Jan. Cureus. 2025. PMID: 39958058 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

