A genetically encoded fluorescent heme sensor detects free heme in plants
- PMID: 38762898
- PMCID: PMC11444292
- DOI: 10.1093/plphys/kiae291
A genetically encoded fluorescent heme sensor detects free heme in plants
Abstract
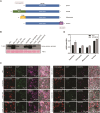
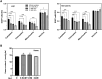
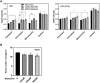
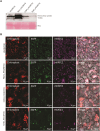
Heme is produced in plants via a plastid-localized metabolic pathway and is subsequently distributed to all cellular compartments. In addition to covalently and noncovalently bound heme, a comparatively small amount of free heme that is not associated with protein is available for incorporation into heme-dependent proteins in all subcellular compartments and for regulatory purposes. This "labile" fraction may also be toxic. To date, the distribution of the free heme pool in plant cells remains poorly understood. Several fluorescence-based methods for the quantification of intracellular free heme have been described. For this study, we used the previously described genetically encoded heme sensor 1 (HS1) to measure the relative amounts of heme in different plant subcellular compartments. In a proof of concept, we manipulated heme content using a range of biochemical and genetic approaches and verified the utility of HS1 in different cellular compartments of Arabidopsis (Arabidopsis thaliana) and tobacco (Nicotiana tabacum and Nicotiana benthamiana) plants transformed either transiently or stably with HS1 and HS1(M7A), a variant with lower affinity for heme. This approach makes it possible to trace the distribution and dynamics of free heme and provides relevant information about its mobilization. The application of these heme sensors will create opportunities to explore and validate the importance of free heme in plant cells and to identify mutants that alter the subcellular allocation of free heme.
© The Author(s) 2024. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Conflict of interest statement
Conflict of interest statement. None declared.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

