CCAT1 lncRNA is chromatin-retained and post-transcriptionally spliced
- PMID: 38763947
- PMCID: PMC11227459
- DOI: 10.1007/s00418-024-02294-w
CCAT1 lncRNA is chromatin-retained and post-transcriptionally spliced
Abstract
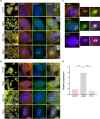
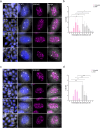
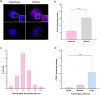
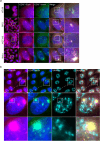
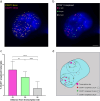
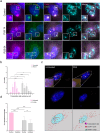
Super-enhancers are unique gene expression regulators widely involved in cancer development. Spread over large DNA segments, they tend to be found next to oncogenes. The super-enhancer c-MYC locus forms long-range chromatin looping with nearby genes, which brings the enhancer and the genes into proximity, to promote gene activation. The colon cancer-associated transcript 1 (CCAT1) gene, which is part of the MYC locus, transcribes a lncRNA that is overexpressed in colon cancer cells through activation by MYC. Comparing different types of cancer cell lines using RNA fluorescence in situ hybridization (RNA FISH), we detected very prominent CCAT1 expression in HeLa cells, observed as several large CCAT1 nuclear foci. We found that dozens of CCAT1 transcripts accumulate on the gene locus, in addition to active transcription occurring from the gene. The accumulating transcripts are released from the chromatin during cell division. Examination of CCAT1 lncRNA expression patterns on the single-RNA level showed that unspliced CCAT1 transcripts are released from the gene into the nucleoplasm. Most of these unspliced transcripts were observed in proximity to the active gene but were not associated with nuclear speckles in which unspliced RNAs usually accumulate. At larger distances from the gene, the CCAT1 transcripts appeared spliced, implying that most CCAT1 transcripts undergo post-transcriptional splicing in the zone of the active gene. Finally, we show that unspliced CCAT1 transcripts can be detected in the cytoplasm during splicing inhibition, which suggests that there are several CCAT1 variants, spliced and unspliced, that the cell can recognize as suitable for export.
Keywords: CCAT1; MYC; PVT1; Post-transcriptional splicing; RNA FISH; Transcription site.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Ahmadiyeh N, Pomerantz MM, Grisanzio C, Herman P, Jia L, Almendro V, He HH, Brown M, Liu XS, Davis M, Caswell JL, Beckwith CA, Hills A, Macconaill L, Coetzee GA, Regan MM, Freedman ML. 8q24 prostate, breast, and colon cancer risk loci show tissue-specific long-range interaction with MYC. Proc Natl Acad Sci U S A. 2010;107(21):9742–9746. doi: 10.1073/pnas.0910668107. - DOI - PMC - PubMed
-
- Alaiyan B, Ilyayev N, Stojadinovic A, Izadjoo M, Roistacher M, Pavlov V, Tzivin V, Halle D, Pan H, Trink B, Gure AO, Nissan A. Differential expression of colon cancer associated transcript1 (CCAT1) along the colonic adenoma-carcinoma sequence. BMC Cancer. 2013;13:196. doi: 10.1186/1471-2407-13-196. - DOI - PMC - PubMed
-
- Barutcu AR, Wu M, Braunschweig U, Dyakov BJA, Luo Z, Turner KM, Durbic T, Lin ZY, Weatheritt RJ, Maass PG, Gingras AC, Blencowe BJ. Systematic mapping of nuclear domain-associated transcripts reveals speckles and lamina as hubs of functionally distinct retained introns. Mol Cell. 2022;82(5):1035–1052 e1039. doi: 10.1016/j.molcel.2021.12.010. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

