Recent Advances in the Discovery of CK2 Inhibitors
- PMID: 38764653
- PMCID: PMC11097362
- DOI: 10.1021/acsomega.3c10478
Recent Advances in the Discovery of CK2 Inhibitors
Abstract
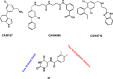
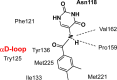
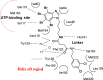
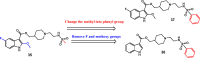
CK2 is a vital enzyme that phosphorylates a large number of substrates and thereby controls many processes in the body. Its upregulation was reported in many cancer types. Inhibitors of CK2 might have anticancer activity, and two compounds are currently under clinical trials. However, both compounds are ATP-competitive inhibitors that may have off-target side effects. The development of allosteric and dual inhibitors can overcome this drawback. These inhibitors showed higher selectivity and specificity for the CK2 enzyme compared to the ATP-competitive inhibitors. The present review summarizes the efforts exerted in the last five years in the design of CK2 inhibitors.
© 2024 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
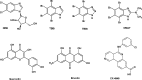
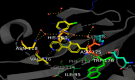

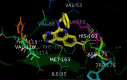
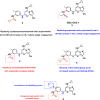

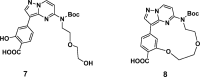

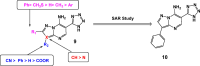
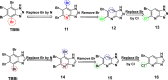
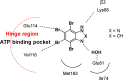








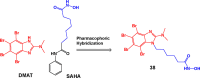
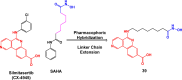

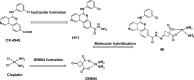



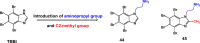

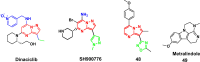
References
-
- Poletto G.; Vilardell J.; Marin O.; Pagano M. A.; Cozza G.; Sarno S.; Itarte E.; Pinna L. A.; Meggio F.; Falques A. The Regulatory Subunit of Protein Kinase CK2 Contributes to the Recognition of the Substrate Consensus Sequence. A Study with an EIF2 -Derived Peptide. Biochemistry 2008, 47, 8317–8325. 10.1021/bi800216d. - DOI - PubMed
-
- Franchin C.; Borgo C.; Cesaro L.; Zaramella S.; Vilardell J.; Salvi M.; Arrigoni G.; Pinna L. A. Re-Evaluation of Protein Kinase CK2 Pleiotropy: New Insights Provided by a Phosphoproteomics Analysis of CK2 Knockout Cells. Cell. Mol. Life Sci. 2018, 75 (11), 2011–2026. 10.1007/s00018-017-2705-8. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
