Overexpression of RBM15 modulated the effect of trophoblast cells by promoting the binding ability between YTHDF2 and the CD82 3'UTR to decrease the expression of CD82
- PMID: 38765115
- PMCID: PMC11098837
- DOI: 10.1016/j.heliyon.2024.e30702
Overexpression of RBM15 modulated the effect of trophoblast cells by promoting the binding ability between YTHDF2 and the CD82 3'UTR to decrease the expression of CD82
Abstract
Background: Pre-eclampsia (PE) is a syndrome with no specific pathological mechanism and is specific to pregnancy. The combined analysis of proteomics and transcriptomics possesses many benefits for treating this disease. m6A modification plays a major role in PE; however, mechanism have not been studied clearly. This study investigated the potential mechanism underlying the role of m6A in PE.
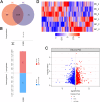
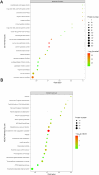
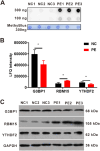
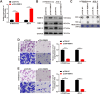
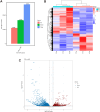
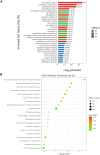
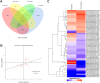
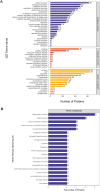
Methods: Mass spectrometry-based label-free quantitative proteomics and transcriptomics experiments were conducted on the placenta of patients with pre-eclampsia and normal pregnancies, and the two omics were followed by joint analysis. Total m6A modification in placental tissues, HTR8/SVneo cells, and JEG-3 cells was measured by dot blot. The levels of RBM15 and CD82 in tissues and cells were detected using qPCR. The protein levels of G3BP1, RBM15, MMP-2, YTHDF2, and MMP-9 were measured by western blotting. The function, migration, and invasion characteristics of HTR8/SVneo and JEG-3 cells were measured using Transwell assays. SRAMP predicted the m6A modification site in the CD82 mRNA 3'UTR, and this was confirmed using luciferase activity and YTHDF2-RIP.
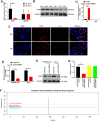
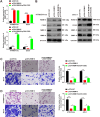
Results: m6A modification was promoted in the PE group, and the RBM15 abundance was increased. Overexpression of RBM15 increased m6A modification. However, overexpression of RBM15 suppressed the expression of MMP-2 and MMP-9 and also the migratory and invasive capabilities of HTR8/SVneo and JEG-3 cells. CD82 expression levels were decreased in PE, and CD82 expression was confirmed via qPCR, western blotting and immunofluorescence. Furthermore, RBM15 overexpression reduced CD82 mRNA and protein levels. Luciferase activity and YTHDF2-RIP results verified that overexpression of RBM15 promoted the binding ability between YTHDF2 and the CD82 3'UTR, thereby decreasing CD82 expression. Finally, CD82 overexpression reversed the effect of RBM15 overexpression on the expression of MMP-2 and MMP-9 and on the migratory and invasive capabilities of the cells.
Conclusions: Overexpression of RBM15 hindered the migratory and invasive capabilities of trophoblasts, while concurrently enhancing m6A modification. The potential mechanism was that overexpression of RBM15 promoted the binding capability between YTHDF2 and CD82 3'UTR and decrease the expression of CD82. Thus, this study provides a theoretical basis for the treatment of PE.
© 2024 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
References
LinkOut - more resources
Full Text Sources
Miscellaneous

