This is a preprint.
Transient interdomain interactions modulate the monomeric structural ensemble and self-assembly of Huntingtin Exon 1
- PMID: 38766024
- PMCID: PMC11100600
- DOI: 10.1101/2024.05.03.592468
Transient interdomain interactions modulate the monomeric structural ensemble and self-assembly of Huntingtin Exon 1
Update in
-
Transient Interdomain Interactions Modulate the Monomeric Structural Ensemble and Self-Assembly of Huntingtin Exon 1.Adv Sci (Weinh). 2025 Jul;12(27):e2501462. doi: 10.1002/advs.202501462. Epub 2025 Apr 28. Adv Sci (Weinh). 2025. PMID: 40289673 Free PMC article.
Abstract
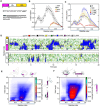
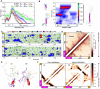
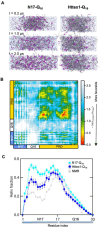
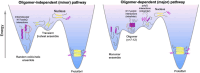
Polyglutamine expansion (≥ 36 residues) within the N-terminal exon-1 of Huntingtin (Httex1) leads to Huntington's disease, a neurodegenerative condition marked by the presence of intranuclear Htt inclusions. Notably, the polyglutamine tract in Httex1 is flanked by an N-terminal coiled-coil domain - N17 (17 amino acids), which undergoes self-association to promote the formation of soluble Httex1 oligomers and brings the aggregation-prone polyQ tracts in close spatial proximity. However, the mechanisms underlying the subsequent conversion of soluble oligomers into insoluble β-rich aggregates with increasing polyQ length, remain unclear. Current knowledge suggests that expansion of the polyQ tract increases its helicity, and this favors its oligomerization and aggregation. In addition, studies utilizing photocrosslinking, conformation-specific antibodies and a stable coiled-coil heterotetrametric system fused to polyQ indicate that domain "cross-talk" (i.e., interdomain interactions) may play a role in the emergence of toxic conformations and the conversion of Httex1 oligomers into fibrillar aggregates. Here, we performed extensive atomistic molecular dynamics (MD) simulations (aggregate time ~ 0.7 ms) to uncover the interplay between structural transformation and domain "cross-talk" on the conformational ensemble and oligomerization landscape of Httex1. Notably, our MD-derived ensembles of N17-polyQ monomers validated against 13C NMR chemical shifts indicated that in addition to elevated α-helicity, polyQ expansion also favors transient, interdomain (N17-polyQ) interactions which result in the emergence of β-sheet conformations. Further, interdomain interactions competed with increased polyQ tract α-helicity to modulate the stability of N17-mediated dimers and thereby promoted a heterogenous dimerization landscape. Finally, we observed that the C-terminal proline-rich domain (PRD) promoted condensation of Httex1 through self-interactions involving its P10/P11 tracts while also interacting with N17 to suppress its α-helicity. In summary, our study demonstrates a significant role for domain "cross-talk" in modulating the monomeric structural ensemble and self-assembly of Httex1.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
