This is a preprint.
The molecular mechanism of on-demand sterol biosynthesis at organelle contact sites
- PMID: 38766039
- PMCID: PMC11100823
- DOI: 10.1101/2024.05.09.593285
The molecular mechanism of on-demand sterol biosynthesis at organelle contact sites
Abstract
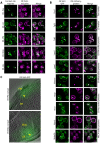
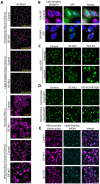
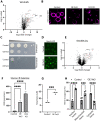
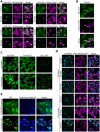
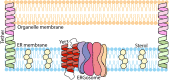
Contact-sites are specialized zones of proximity between two organelles, essential for organelle communication and coordination. The formation of contacts between the Endoplasmic Reticulum (ER), and other organelles, relies on a unique membrane environment enriched in sterols. However, how these sterol-rich domains are formed and maintained had not been understood. We found that the yeast membrane protein Yet3, the homolog of human BAP31, is localized to multiple ER contact sites. We show that Yet3 interacts with all the enzymes of the post-squalene ergosterol biosynthesis pathway and recruits them to create sterol-rich domains. Increasing sterol levels at ER contacts causes its depletion from the plasma membrane leading to a compensatory reaction and altered cell metabolism. Our data shows that Yet3 provides on-demand sterols at contacts thus shaping organellar structure and function. A molecular understanding of this protein's functions gives new insights into the role of BAP31 in development and pathology.
Figures
Similar articles
-
Endoplasmic reticulum-plasma membrane contact sites integrate sterol and phospholipid regulation.PLoS Biol. 2018 May 21;16(5):e2003864. doi: 10.1371/journal.pbio.2003864. eCollection 2018 May. PLoS Biol. 2018. PMID: 29782498 Free PMC article.
-
Ergosterol interacts with Sey1p to promote atlastin-mediated endoplasmic reticulum membrane fusion in Saccharomyces cerevisiae.FASEB J. 2019 Mar;33(3):3590-3600. doi: 10.1096/fj.201800779RR. Epub 2018 Nov 21. FASEB J. 2019. PMID: 30462528
-
Structural basis of sterol recognition and nonvesicular transport by lipid transfer proteins anchored at membrane contact sites.Proc Natl Acad Sci U S A. 2018 Jan 30;115(5):E856-E865. doi: 10.1073/pnas.1719709115. Epub 2018 Jan 16. Proc Natl Acad Sci U S A. 2018. PMID: 29339490 Free PMC article.
-
Mechanisms of sterol uptake and transport in yeast.J Steroid Biochem Mol Biol. 2012 Mar;129(1-2):70-8. doi: 10.1016/j.jsbmb.2010.11.014. Epub 2010 Dec 8. J Steroid Biochem Mol Biol. 2012. PMID: 21145395 Review.
-
Sterol trafficking between the endoplasmic reticulum and plasma membrane in yeast.Biochem Soc Trans. 2006 Jun;34(Pt 3):356-8. doi: 10.1042/BST0340356. Biochem Soc Trans. 2006. PMID: 16709160 Review.
Cited by
-
Nondisruptive inducible labeling of ER-membrane contact sites using the Lamin B receptor.PLoS Biol. 2025 Jul 10;23(7):e3003249. doi: 10.1371/journal.pbio.3003249. eCollection 2025 Jul. PLoS Biol. 2025. PMID: 40638559 Free PMC article.
References
-
- Shai N., Yifrach E., Van Roermund C.W.T., Cohen N., Bibi C., Ijlst L., Cavellini L., Meurisse J., Schuster R., Zada L., et al. (2018). Systematic mapping of contact sites reveals tethers and a function for the peroxisome-mitochondria contact. Nat Commun 9. 10.1038/s41467-018-03957-8. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
