This is a preprint.
Bayesian Estimation of Muscle Mechanisms and Therapeutic Targets Using Variational Autoencoders
- PMID: 38766103
- PMCID: PMC11100674
- DOI: 10.1101/2024.05.08.593035
Bayesian Estimation of Muscle Mechanisms and Therapeutic Targets Using Variational Autoencoders
Update in
-
Bayesian estimation of muscle mechanisms and therapeutic targets using variational autoencoders.Biophys J. 2025 Jan 7;124(1):179-191. doi: 10.1016/j.bpj.2024.11.3310. Epub 2024 Nov 26. Biophys J. 2025. PMID: 39604261 Free PMC article.
Abstract
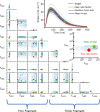
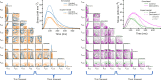
Cardiomyopathies, often caused by mutations in genes encoding muscle proteins, are traditionally treated by phenotyping hearts and addressing symptoms post irreversible damage. With advancements in genotyping, early diagnosis is now possible, potentially introducing earlier treatment. However, the intricate structure of muscle and its myriad proteins make treatment predictions challenging. Here we approach the problem of estimating therapeutic targets for a mutation in mouse muscle using a spatially explicit half sarcomere muscle model. We selected 9 rate parameters in our model linked to both small molecules and cardiomyopathy-causing mutations. We then randomly varied these rate parameters and simulated an isometric twitch for each combination to generate a large training dataset. We used this dataset to train a Conditional Variational Autoencoder (CVAE), a technique used in Bayesian parameter estimation. Given simulated or experimental isometric twitches, this machine learning model is able to then predict the set of rate parameters which are most likely to yield that result. We then predict the set of rate parameters associated with twitches from control mice with the cardiac Troponin C (cTnC) I61Q variant and control twitches treated with the myosin activator Danicamtiv, as well as model parameters that recover the abnormal I61Q cTnC twitches.
Conflict of interest statement
DECLARATION OF INTERESTS The authors declare no competing interests.
Figures






References
-
- Maughan D. W., and Vigoreaux J. O., 1999. An Integrated View of Insect Flight Muscle: Genes, Motor Molecules, and Motion. Physiology 14:87–92. - PubMed
Publication types
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
