This is a preprint.
Conformational switching of Arp5 subunit differentially regulates INO80 chromatin remodeling
- PMID: 38766108
- PMCID: PMC11100795
- DOI: 10.1101/2024.05.10.593625
Conformational switching of Arp5 subunit differentially regulates INO80 chromatin remodeling
Update in
-
Conformational switching of Arp5 subunit regulates INO80 chromatin remodeling.Nucleic Acids Res. 2025 Jan 11;53(2):gkae1187. doi: 10.1093/nar/gkae1187. Nucleic Acids Res. 2025. PMID: 39676660 Free PMC article.
Abstract
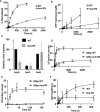
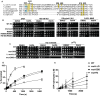
The INO80 chromatin remodeler is a versatile enzyme capable of several functions, including spacing nucleosomes equal distances apart, precise positioning of nucleosomes based on DNA shape/sequence and exchanging histone dimers. Within INO80, the Arp5 subunit plays a central role in INO80 remodeling, evidenced by its interactions with the histone octamer, nucleosomal and extranucleosomal DNA, and its necessity in linking INO80's ATPase activity to nucleosome movement. Our investigation reveals that the grappler domain of Arp5 interacts with the acidic pocket of nucleosomes through two distinct mechanisms: an arginine anchor or a hydrophobic/acidic patch. These two modes of binding serve distinct functions within INO80 as shown in vivo by mutations in these regions resulting in varying phenotypes and in vitro by diverse effects on nucleosome mobilization. Our findings suggest that the hydrophobic/acidic patch of Arp5 is likely important for dimer exchange by INO80, while the arginine anchor is crucial for mobilizing nucleosomes.
Keywords: ARP5; DNA repair; DNA replication stress; INO80; INO80 structure; chromatin/nucleosome remodeling; nucleosome dynamics.
Conflict of interest statement
Conflict of Interests The authors declare no competing interests.
Figures





References
-
- Sureshkumar S. and Balasubramanian S. (2021) Complexes and complexities: INO80 takes center stage. Mol Plant, 14, 1776–1778. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
