This is a preprint.
Excitatory Neuron-Derived Interleukin-34 Controls Cortical Developmental Microglia Function
- PMID: 38766127
- PMCID: PMC11100801
- DOI: 10.1101/2024.05.10.589920
Excitatory Neuron-Derived Interleukin-34 Controls Cortical Developmental Microglia Function
Update in
-
Excitatory-neuron-derived interleukin-34 supports cortical developmental microglia function.Immunity. 2025 Aug 12;58(8):1948-1965.e6. doi: 10.1016/j.immuni.2025.06.002. Epub 2025 Jul 2. Immunity. 2025. PMID: 40609535
Abstract
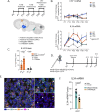
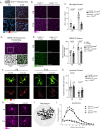
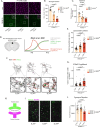
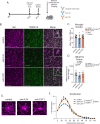
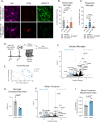
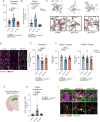
Neuron-microglia interactions dictate the development of neuronal circuits in the brain. However, the factors that regulate these processes across development are largely unknown. Here, we find that IL34, a neuron-derived cytokine, is upregulated in early development and maintains neuroprotective, mature microglia in the anterior cingulate cortex (ACC) of mice. We show that IL34 is upregulated in the second week of postnatal life and is expressed primarily in excitatory neurons. Excitatory-neuron specific knock-out of IL34 reduced microglia number and TMEM119 expression and increased aberrant microglial phagocytosis of excitatory thalamocortical synapses in the ACC. Acute, low dose blocking of IL34 at postnatal day 15 similarly decreased TMEM119 and inappropriately increased microglial phagocytosis of synapses. Viral overexpression of IL34 induced TMEM119 expression and prevented appropriate microglial phagocytosis of synapses. These findings establish IL34 as a key regulator of neuron-microglia crosstalk in postnatal brain development, controlling both microglial maturation and synapse engulfment.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures

References
-
- Nandi S., Gokhan S., Dai X.-M., Wei S., Enikolopov G., Lin H., Mehler M.F., and Stanley E.R. (2012). The CSF-1 receptor ligands IL-34 and CSF-1 exhibit distinct developmental brain expression patterns and regulate neural progenitor cell maintenance and maturation. Dev. Biol. 367, 100–113. 10.1016/j.ydbio.2012.03.026. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
