This is a preprint.
Asesino: a nucleus-forming phage that lacks PhuZ
- PMID: 38766163
- PMCID: PMC11100802
- DOI: 10.1101/2024.05.10.593592
Asesino: a nucleus-forming phage that lacks PhuZ
Update in
-
Erwinia phage Asesino is a nucleus-forming phage that lacks PhuZ.Sci Rep. 2025 Jan 11;15(1):1692. doi: 10.1038/s41598-024-64095-4. Sci Rep. 2025. PMID: 39799172 Free PMC article.
Abstract
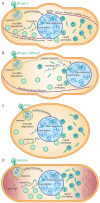
As nucleus-forming phages become better characterized, understanding their unifying similarities and unique differences will help us understand how they occupy varied niches and infect diverse hosts. All identified nucleus-forming phages fall within the proposed Chimalliviridae family and share a core genome of 68 unique genes including chimallin, the major nuclear shell protein. A well-studied but non-essential protein encoded by many nucleus-forming phages is PhuZ, a tubulin homolog which aids in capsid migration, nucleus rotation, and nucleus positioning. One clade that represents 24% of all currently known chimalliviruses lacks a PhuZ homolog. Here we show that Erwinia phage Asesino, one member of this PhuZ-less clade, shares a common overall replication mechanism with other characterized nucleus-forming phages despite lacking PhuZ. We show that Asesino replicates via a phage nucleus that encloses phage DNA and partitions proteins in the nuclear compartment and cytoplasm in a manner similar to previously characterized nucleus-forming phages. Consistent with a lack of PhuZ, however, we did not observe active positioning or rotation of the phage nucleus within infected cells. These data show that some nucleus-forming phages have evolved to replicate efficiently without PhuZ, providing an example of a unique variation in the nucleus-based replication pathway.
Conflict of interest statement
COMPETING INTERESTS STATEMENT J.P. has an equity interest in Linnaeus Bioscience Incorporated and receives income. The terms of this arrangement have been reviewed and approved by the University of California, San Diego, in accordance with its conflict-of-interest policies.
Figures




References
-
- Malone L. M. et al. A jumbo phage that forms a nucleus-like structure evades CRISPR-Cas DNA targeting but is vulnerable to type III RNA-based immunity. Nat Microbiol 5, 48–55 (2020). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
