This is a preprint.
Functional interrogation of cellular Lp(a) uptake by genome-scale CRISPR screening
- PMID: 38766193
- PMCID: PMC11100788
- DOI: 10.1101/2024.05.11.593568
Functional interrogation of cellular Lp(a) uptake by genome-scale CRISPR screening
Update in
-
Functional interrogation of cellular Lp(a) uptake by genome-scale CRISPR screening.Atherosclerosis. 2025 Apr;403:119174. doi: 10.1016/j.atherosclerosis.2025.119174. Epub 2025 Mar 22. Atherosclerosis. 2025. PMID: 40174266
Abstract
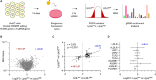
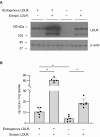
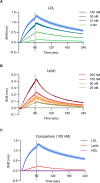
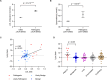
An elevated level of lipoprotein(a), or Lp(a), in the bloodstream has been causally linked to the development of atherosclerotic cardiovascular disease and calcific aortic valve stenosis. Steady state levels of circulating lipoproteins are modulated by their rate of clearance, but the identity of the Lp(a) uptake receptor(s) has been controversial. In this study, we performed a genome-scale CRISPR screen to functionally interrogate all potential Lp(a) uptake regulators in HuH7 cells. Strikingly, the top positive and negative regulators of Lp(a) uptake in our screen were LDLR and MYLIP, encoding the LDL receptor and its ubiquitin ligase IDOL, respectively. We also found a significant correlation for other genes with established roles in LDLR regulation. No other gene products, including those previously proposed as Lp(a) receptors, exhibited a significant effect on Lp(a) uptake in our screen. We validated the functional influence of LDLR expression on HuH7 Lp(a) uptake, confirmed in vitro binding between the LDLR extracellular domain and purified Lp(a), and detected an association between loss-of-function LDLR variants and increased circulating Lp(a) levels in the UK Biobank cohort. Together, our findings support a central role for the LDL receptor in mediating Lp(a) uptake by hepatocytes.
Conflict of interest statement
Declaration of interests The authors have no relevant competing financial interests to declare.
Figures

References
-
- Seman L. J. et al. Lipoprotein(a)-cholesterol and coronary heart disease in the Framingham Heart Study. Clin Chem 45, 1039–1046 (1999). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
