This is a preprint.
Microbiota and metabolic adaptation shape Staphylococcus aureus virulence and antimicrobial resistance during intestinal colonization
- PMID: 38766195
- PMCID: PMC11100824
- DOI: 10.1101/2024.05.11.593044
Microbiota and metabolic adaptation shape Staphylococcus aureus virulence and antimicrobial resistance during intestinal colonization
Abstract
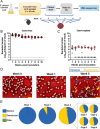
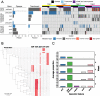
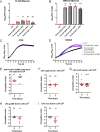
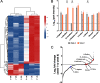
Depletion of microbiota increases susceptibility to gastrointestinal colonization and subsequent infection by opportunistic pathogens such as methicillin-resistant Staphylococcus aureus (MRSA). How the absence of gut microbiota impacts the evolution of MRSA is unknown. The present report used germ-free mice to investigate the evolutionary dynamics of MRSA in the absence of gut microbiota. Through genomic analyses and competition assays, we found that MRSA adapts to the microbiota-free gut through sequential genetic mutations and structural changes that enhance fitness. Initially, these adaptations increase carbohydrate transport; subsequently, evolutionary pathways largely diverge to enhance either arginine metabolism or cell wall biosynthesis. Increased fitness in arginine pathway mutants depended on arginine catabolic genes, especially nos and arcC, which promote microaerobic respiration and ATP generation, respectively. Thus, arginine adaptation likely improves redox balance and energy production in the oxygen-limited gut environment. Findings were supported by human gut metagenomic analyses, which suggest the influence of arginine metabolism on colonization. Surprisingly, these adaptive genetic changes often reduced MRSA's antimicrobial resistance and virulence. Furthermore, resistance mutation, typically associated with decreased virulence, also reduced colonization fitness, indicating evolutionary trade-offs among these traits. The presence of normal microbiota inhibited these adaptations, preserving MRSA's wild-type characteristics that effectively balance virulence, resistance, and colonization fitness. The results highlight the protective role of gut microbiota in preserving a balance of key MRSA traits for long-term ecological success in commensal populations, underscoring the potential consequences on MRSA's survival and fitness during and after host hospitalization and antimicrobial treatment.
Conflict of interest statement
CONFLICTS OF INTERESTS B.S. has consulted for Basilea Pharmaceutica. V.J.T. has received honoraria from Pfizer and MedImmune and is an inventor on patents and patent applications filed by New York University, which are currently under commercial license to Janssen Biotech Inc. Janssen Biotech Inc. provides research funding and other payments associated with a licensing agreement. K.C. has received research support from Pfizer, Takeda, Pacific Biosciences, Genentech, and AbbVie, consulted for or received honoraria from Vedanta, Genentech, and AbbVie, and is an inventor on US patent 10,722,600 and pro- visional patents 62/935,035 and 63/157,225. J.S. holds equity in Postbiotics Plus Research, has filed intellectual property applications related to the microbiome (reference numbers #63/299,607), and is on an advisory board and holds equity of Jona Health.
Figures


References
-
- von Eiff C, Becker K, Machka K, Stammer H, Peters G. 2001. Nasal carriage as a source of Staphylococcus aureus bacteremia. Study Group. N Engl J Med 344:11–6. - PubMed
-
- Wertheim HF, Melles DC, Vos MC, van Leeuwen W, van Belkum A, Verbrugh HA, Nouwen JL. 2005. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis 5:751–62. - PubMed
-
- Faden H, Lesse AJ, Trask J, Hill JA, Hess DJ, Dryja D, Lee YH. 2010. Importance of colonization site in the current epidemic of staphylococcal skin abscesses. Pediatrics 125:e618–24. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
