Mycobacterial CpsA activates type I IFN signaling in macrophages via cGAS-mediated pathway
- PMID: 38766355
- PMCID: PMC11099328
- DOI: 10.1016/j.isci.2024.109807
Mycobacterial CpsA activates type I IFN signaling in macrophages via cGAS-mediated pathway
Abstract
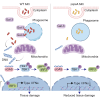
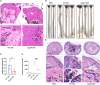
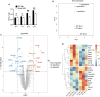
Type I interferon (IFN) production is crucial in tuberculosis pathogenesis, yet the bacterial factors initiating this process are incompletely understood. CpsA, protein of Mycobacterium marinum and Mycobacterium tuberculosis, plays a key role in maintaining bacterial virulence and inhibiting host cell LC3-associated phagocytosis. By utilizing CpsA full deletion mutant studies, we re-verified its essential role in infection-induced pathology and revealed its new role in type I IFN expression. CpsA deficiency hindered IFN production in infected macrophages in vitro as well as zebrafish and mice in vivo. This effect was linked to the cGAS-TBK1-IRF3 pathway, as evidenced by decreased TBK1 and IRF3 phosphorylation in CpsA-deficient bacterial strain-infected macrophages. Moreover, we further show that CpsA deficiency cause decreased cytosolic DNA levels, correlating with impaired phagosomal membrane rupture. Our findings reveal a new function of mycobacterial CpsA in type I IFN production and offer insight into the molecular mechanisms underlying mycobacterial infection pathology.
Keywords: Immunology; Microbiology; Molecular microbiology.
© 2024 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Teles R.M.B., Graeber T.G., Krutzik S.R., Montoya D., Schenk M., Lee D.J., Komisopoulou E., Kelly-Scumpia K., Chun R., Iyer S.S., et al. Type I interferon suppresses type II interferon-triggered human anti-mycobacterial responses. Science. 2013;339:1448–1453. doi: 10.1126/science.1233665. - DOI - PMC - PubMed
-
- Berry M.P.R., Graham C.M., McNab F.W., Xu Z., Bloch S.A.A., Oni T., Wilkinson K.A., Banchereau R., Skinner J., Wilkinson R.J., et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466:973–977. doi: 10.1038/nature09247. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

