Zinc finger knuckle genes are associated with tolerance to drought and dehydration in chickpea (Cicer arietinum L.)
- PMID: 38766473
- PMCID: PMC11099236
- DOI: 10.3389/fpls.2024.1354413
Zinc finger knuckle genes are associated with tolerance to drought and dehydration in chickpea (Cicer arietinum L.)
Abstract
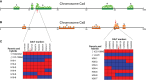
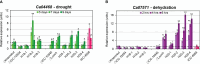
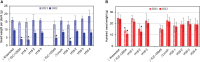
Chickpea (Cicer arietinum L.) is a very important food legume and needs improved drought tolerance for higher seed production in dry environments. The aim of this study was to determine diversity and genetic polymorphism in zinc finger knuckle genes with CCHC domains and their functional analysis for practical improvement of chickpea breeding. Two CaZF-CCHC genes, Ca04468 and Ca07571, were identified as potentially important candidates associated with plant responses to drought and dehydration. To study these genes, various methods were used including Sanger sequencing, DArT (Diversity array technology) and molecular markers for plant genotyping, gene expression analysis using RT-qPCR, and associations with seed-related traits in chickpea plants grown in field trials. These genes were studied for genetic polymorphism among a set of chickpea accessions, and one SNP was selected for further study from four identified SNPs between the promoter regions of each of the two genes. Molecular markers were developed for the SNP and verified using the ASQ and CAPS methods. Genotyping of parents and selected breeding lines from two hybrid populations, and SNP positions on chromosomes with haplotype identification, were confirmed using DArT microarray analysis. Differential expression profiles were identified in the parents and the hybrid populations under gradual drought and rapid dehydration. The SNP-based genotypes were differentially associated with seed weight per plant but not with 100 seed weight. The two developed and verified SNP molecular markers for both genes, Ca04468 and Ca07571, respectively, could be used for marker-assisted selection in novel chickpea cultivars with improved tolerance to drought and dehydration.
Keywords: CCHC domain; DArT analysis; SNP; chickpea; drought and dehydration; gene expression; seed yield; zinc finger knuckle gene.
Copyright © 2024 Khassanova, Oshergina, Ten, Jatayev, Zhanbyrshina, Gabdola, Gupta, Schramm, Pupulin, Philp-Dutton, Anderson, Sweetman, Jenkins, Soole and Shavrukov.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

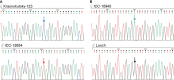
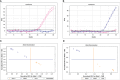
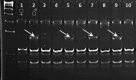
References
-
- Aklilu E. (2021). Review on forward and reverse genetics in plant breeding. All Life 14, 127–135. doi: 10.1080/26895293.2021.1888810 - DOI
-
- Amangeldiyeva A., Baidyussen A., Kuzbakova M., Yerzhebayeva R., Jatayev S., Shavrukov Y. (2023). “Modified allele-specific qPCR (ASQ) genotyping,” in Plant Genotyping: Methods and Protocols. Methods in Molecular Biology, vol. 2638 . Ed. Shavrukov Y. (SpringerNature, New York: ), 231–247. doi: 10.1007/978-1-0716-3024-2_16 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

