Alkenyl oxindole is a novel PROTAC moiety that recruits the CRL4DCAF11 E3 ubiquitin ligase complex for targeted protein degradation
- PMID: 38768083
- PMCID: PMC11104598
- DOI: 10.1371/journal.pbio.3002550
Alkenyl oxindole is a novel PROTAC moiety that recruits the CRL4DCAF11 E3 ubiquitin ligase complex for targeted protein degradation
Abstract
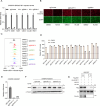
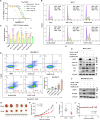
Alkenyl oxindoles have been characterized as autophagosome-tethering compounds (ATTECs), which can target mutant huntingtin protein (mHTT) for lysosomal degradation. In order to expand the application of alkenyl oxindoles for targeted protein degradation, we designed and synthesized a series of heterobifunctional compounds by conjugating different alkenyl oxindoles with bromodomain-containing protein 4 (BRD4) inhibitor JQ1. Through structure-activity relationship study, we successfully developed JQ1-alkenyl oxindole conjugates that potently degrade BRD4. Unexpectedly, we found that these molecules degrade BRD4 through the ubiquitin-proteasome system, rather than the autophagy-lysosomal pathway. Using pooled CRISPR interference (CRISPRi) screening, we revealed that JQ1-alkenyl oxindole conjugates recruit the E3 ubiquitin ligase complex CRL4DCAF11 for substrate degradation. Furthermore, we validated the most potent heterobifunctional molecule HL435 as a promising drug-like lead compound to exert antitumor activity both in vitro and in a mouse xenograft tumor model. Our research provides new employable proteolysis targeting chimera (PROTAC) moieties for targeted protein degradation, providing new possibilities for drug discovery.
Copyright: © 2024 Wang et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Ciechanover A. Intracellular Protein Degradation: From a Vague Idea, through the Lysosome and the Ubiquitin–Proteasome System, and onto Human Diseases and Drug Targeting. Angew Chem Int Ed. 2005;44(37):5944–5967. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

