Genome-wide screening of meta-QTL and candidate genes controlling yield and yield-related traits in barley (Hordeum vulgare L.)
- PMID: 38768114
- PMCID: PMC11104655
- DOI: 10.1371/journal.pone.0303751
Genome-wide screening of meta-QTL and candidate genes controlling yield and yield-related traits in barley (Hordeum vulgare L.)
Abstract
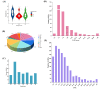
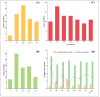
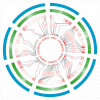
Increasing yield is an important goal of barley breeding. In this study, 54 papers published from 2001-2022 on QTL mapping for yield and yield-related traits in barley were collected, which contained 1080 QTLs mapped to the barley high-density consensus map for QTL meta-analysis. These initial QTLs were integrated into 85 meta-QTLs (MQTL) with a mean confidence interval (CI) of 2.76 cM, which was 7.86-fold narrower than the CI of the initial QTL. Among these 85 MQTLs, 68 MQTLs were validated in GWAS studies, and 25 breeder's MQTLs were screened from them. Seventeen barley orthologs of yield-related genes in rice and maize were identified within the hcMQTL region based on comparative genomics strategy and were presumed to be reliable candidates for controlling yield-related traits. The results of this study provide useful information for molecular marker-assisted breeding and candidate gene mining of yield-related traits in barley.
Copyright: © 2024 Du et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Genome wide screening and comparative genome analysis for Meta-QTLs, ortho-MQTLs and candidate genes controlling yield and yield-related traits in rice.BMC Genomics. 2020 Apr 10;21(1):294. doi: 10.1186/s12864-020-6702-1. BMC Genomics. 2020. PMID: 32272882 Free PMC article.
-
Detection of consensus genomic regions and candidate genes for quality traits in barley using QTL meta-analysis.Front Plant Sci. 2024 Jan 11;14:1319889. doi: 10.3389/fpls.2023.1319889. eCollection 2023. Front Plant Sci. 2024. PMID: 38283973 Free PMC article.
-
Meta-QTLs, ortho-meta-QTLs and candidate genes for grain yield and associated traits in wheat (Triticum aestivum L.).Theor Appl Genet. 2022 Mar;135(3):1049-1081. doi: 10.1007/s00122-021-04018-3. Epub 2022 Jan 5. Theor Appl Genet. 2022. PMID: 34985537
-
Genetic Localization and Homologous Genes Mining for Barley Grain Size.Int J Mol Sci. 2023 Mar 3;24(5):4932. doi: 10.3390/ijms24054932. Int J Mol Sci. 2023. PMID: 36902360 Free PMC article. Review.
-
Barley2035: A decadal vision for barley research and breeding.Mol Plant. 2025 Feb 3;18(2):195-218. doi: 10.1016/j.molp.2024.12.009. Epub 2024 Dec 17. Mol Plant. 2025. PMID: 39690737 Review.
Cited by
-
Genetic dissection of yield and yield-related traits in mungbean based on QTL meta-analysis.Front Genet. 2025 May 8;16:1600979. doi: 10.3389/fgene.2025.1600979. eCollection 2025. Front Genet. 2025. PMID: 40406059 Free PMC article.
-
New loci and candidate genes in spring two-rowed barley detected through meta-analysis of a field trial European network.Theor Appl Genet. 2025 Jun 23;138(7):158. doi: 10.1007/s00122-025-04934-8. Theor Appl Genet. 2025. PMID: 40549197 Free PMC article.
-
Identification of QTLs associated with grain yield-related traits of spring barley.BMC Plant Biol. 2025 Apr 28;25(1):554. doi: 10.1186/s12870-025-06588-6. BMC Plant Biol. 2025. PMID: 40295941 Free PMC article.
References
-
- Badr A, Müller K, Sch R, Rabey H, Effgen S, Ibrahim H, et al.. On the origin and domestication history of Barley (Hordeum vulgare). Mol Biol Evol. 2000;17: 499–510. - PubMed
-
- Yin X, Chasalow SD, Stam P, Kropff MJ, Dourleijn CJ, Bos I, et al.. Use of component analysis in QTL mapping of complex crop traits: a case study on yield in barley. Plant Breeding. 2002;121: 314–319.
-
- Li JZ, Huang XQ, Heinrichs F, Ganal MW, Roder MS. Analysis of QTLs for yield, yield components, and malting quality in a BC3-DH population of spring barley. Theor Appl Genet. 2005;110: 356–363. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

