Wiring Between Close Nodes in Molecular Networks Evolves More Quickly Than Between Distant Nodes
- PMID: 38768245
- PMCID: PMC11136681
- DOI: 10.1093/molbev/msae098
Wiring Between Close Nodes in Molecular Networks Evolves More Quickly Than Between Distant Nodes
Abstract
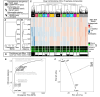
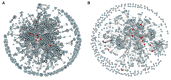
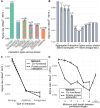
As species diverge, a wide range of evolutionary processes lead to changes in protein-protein interaction (PPI) networks and metabolic networks. The rate at which molecular networks evolve is an important question in evolutionary biology. Previous empirical work has focused on interactomes from model organisms to calculate rewiring rates, but this is limited by the relatively small number of species and sparse nature of network data across species. We present a proxy for variation in network topology: variation in drug-drug interactions (DDIs), obtained by studying drug combinations (DCs) across taxa. Here, we propose the rate at which DDIs change across species as an estimate of the rate at which the underlying molecular network changes as species diverge. We computed the evolutionary rates of DDIs using previously published data from a high-throughput study in gram-negative bacteria. Using phylogenetic comparative methods, we found that DDIs diverge rapidly over short evolutionary time periods, but that divergence saturates over longer time periods. In parallel, we mapped drugs with known targets in PPI and cofunctional networks. We found that the targets of synergistic DDIs are closer in these networks than other types of DCs and that synergistic interactions have a higher evolutionary rate, meaning that nodes that are closer evolve at a faster rate. Future studies of network evolution may use DC data to gain larger-scale perspectives on the details of network evolution within and between species.
Keywords: biological network evolution; drug–drug interactions; gram-negative bacteria; phylogenetic comparative methods.
© The Author(s) 2024. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures

References
-
- Adams DC, Otárola‐Castillo E, Paradis E. geomorph: an R package for the collection and analysis of geometric morphometric shape data. Methods in Ecology and Evolution. 2013:4(4):393–399. 10.1111/2041-210X.12035. - DOI
-
- Armstrong JF, Faccenda E, Harding SD, Pawson AJ, Southan C, Sharman JL, Campo B, Cavanagh DR, Alexander SPH, Davenport AP, et al. The IUPHAR/BPS guide to PHARMACOLOGY in 2020: extending immunopharmacology content and introducing the IUPHAR/MMV guide to MALARIA PHARMACOLOGY. Nucleic Acids Res. 2020:48(D1):D1006–D1021. 10.1093/nar/gkz951. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

