Inhibition of mRNA nuclear export promotes SARS-CoV-2 pathogenesis
- PMID: 38768348
- PMCID: PMC11145185
- DOI: 10.1073/pnas.2314166121
Inhibition of mRNA nuclear export promotes SARS-CoV-2 pathogenesis
Abstract
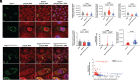
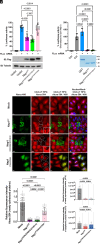
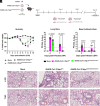
The nonstructural protein 1 (Nsp1) of SARS-CoV-2 (Severe Acute Respiratory Syndrome Coronavirus 2) is a virulence factor that targets multiple cellular pathways to inhibit host gene expression and antiviral response. However, the underlying mechanisms of the various Nsp1-mediated functions and their contributions to SARS-CoV-2 virulence remain unclear. Among the targets of Nsp1 is the mRNA (messenger ribonucleic acid) export receptor NXF1-NXT1, which mediates nuclear export of mRNAs from the nucleus to the cytoplasm. Based on Nsp1 crystal structure, we generated mutants on Nsp1 surfaces and identified an acidic N-terminal patch that is critical for interaction with NXF1-NXT1. Photoactivatable Nsp1 probe reveals the RNA Recognition Motif (RRM) domain of NXF1 as an Nsp1 N-terminal binding site. By mutating the Nsp1 N-terminal acidic patch, we identified a separation-of-function mutant of Nsp1 that retains its translation inhibitory function but substantially loses its interaction with NXF1 and reverts Nsp1-mediated mRNA export inhibition. We then generated a recombinant (r)SARS-CoV-2 mutant on the Nsp1 N-terminal acidic patch and found that this surface is key to promote NXF1 binding and inhibition of host mRNA nuclear export, viral replication, and pathogenicity in vivo. Thus, these findings provide a mechanistic understanding of Nsp1-mediated mRNA export inhibition and establish the importance of this pathway in the virulence of SARS-CoV-2.
Keywords: NXF1; Nsp1; SARS-CoV-2; mRNA export; nuclear transport.
Conflict of interest statement
Competing interests statement:A.G.-S. has consulting agreements for the following companies involving cash and/or stock: Castlevax, Amovir, Vivaldi Biosciences, Contrafect, 7Hills Pharma, Avimex, Pagoda, Accurius, Esperovax, Farmak, Applied Biological Laboratories, Pharmamar, CureLab Oncology, CureLab Veterinary, Synairgen, Paratus, and Pfizer, outside of the reported work. A.G.-S. has been an invited speaker in meeting events organized by Seqirus, Janssen, Abbott, and Astrazeneca. A.G.-S. is inventor on patents and patent applications on the use of antivirals and vaccines for the treatment and prevention of virus infections and cancer, owned by the Icahn School of Medicine at Mount Sinai, New York.
Figures



Comment in
-
SARS-CoV-2 Nsp1 traps RNA in the nucleus to escape immune detection.Proc Natl Acad Sci U S A. 2024 Jun 18;121(25):e2408794121. doi: 10.1073/pnas.2408794121. Epub 2024 Jun 6. Proc Natl Acad Sci U S A. 2024. PMID: 38843251 Free PMC article. No abstract available.
References
-
- Finkel Y., et al. , SARS-CoV-2 uses a multipronged strategy to impede host protein synthesis. Nature 594, 240–245 (2021). - PubMed
MeSH terms
Substances
Grants and funding
- U54 CA260560/CA/NCI NIH HHS/United States
- U19AI171443/HHS | National Institutes of Health (NIH)
- CA260560/HHS | NIH | National Cancer Institute (NCI)
- S10-OD026983/HHS | National Institutes of Health (NIH)
- U19 AI171443/AI/NIAID NIH HHS/United States
- P30 CA142543/CA/NCI NIH HHS/United States
- U19AI135972/HHS | National Institutes of Health (NIH)
- R35 GM133743/GM/NIGMS NIH HHS/United States
- GM133743/HHS | National Institutes of Health (NIH)
- #75N93021C00014/HHS | National Institutes of Health (NIH)
- S10OD030269/HHS | National Institutes of Health (NIH)
- S10 OD026983/OD/NIH HHS/United States
- R01 AI184975/AI/NIAID NIH HHS/United States
- S10 OD030269/OD/NIH HHS/United States
- U19 AI135972/AI/NIAID NIH HHS/United States
- 75N93021C00014/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

