Inflammatory and neurodegenerative serum protein biomarkers increase sensitivity to detect clinical and radiographic disease activity in multiple sclerosis
- PMID: 38769309
- PMCID: PMC11106245
- DOI: 10.1038/s41467-024-48602-9
Inflammatory and neurodegenerative serum protein biomarkers increase sensitivity to detect clinical and radiographic disease activity in multiple sclerosis
Abstract
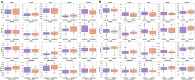
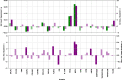
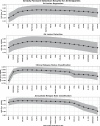
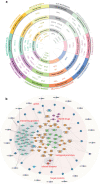
The multifaceted nature of multiple sclerosis requires quantitative biomarkers that can provide insights related to diverse physiological pathways. To this end, proteomic analysis of deeply-phenotyped serum samples, biological pathway modeling, and network analysis were performed to elucidate inflammatory and neurodegenerative processes, identifying sensitive biomarkers of multiple sclerosis disease activity. Here, we evaluated the concentrations of > 1400 serum proteins in 630 samples from three multiple sclerosis cohorts for association with clinical and radiographic new disease activity. Twenty proteins were associated with increased clinical and radiographic multiple sclerosis disease activity for inclusion in a custom assay panel. Serum neurofilament light chain showed the strongest univariate correlation with gadolinium lesion activity, clinical relapse status, and annualized relapse rate. Multivariate modeling outperformed univariate for all endpoints. A comprehensive biomarker panel including the twenty proteins identified in this study could serve to characterize disease activity for a patient with multiple sclerosis.
© 2024. The Author(s).
Conflict of interest statement
The authors declare the following competing interests: T.C. has received compensation for consulting from Biogen, Novartis Pharmaceuticals, Roche Genentech, and Sanofi Genzyme, and received research support from the National Institutes of Health, National MS Society, US Department of Defense, EMD Serono, I-Mab Biopharma, Novartis Pharmaceuticals, Octave Bioscience, Inc, Roche Genentech, and Tiziana Life Sciences. V.M.G., M.B., and A.K. were employees of Octave Bioscience, Inc at the time the study was completed. R.B. is funded by the NMSS Harry Weaver Award, NIH, DOD, NSF, as well as Biogen, Novartis, and Roche Genentech. She has received personal fees for consulting from Alexion, EMD Serono, Horizon, Janssen, Sanofi-Genzyme, and TG Therapeutics. B.A.C.C. has received personal compensation for consulting from Alexion, Atara, Autobahn, Avotres, Biogen, Boston Pharma, EMD Serono, Gossamer Bio, Hexal/Sandoz, Horizon, Immunic AG, Neuron23, Novartis, Sanofi, Siemens, and TG Therapeutics, and received research support from Genentech. S.L.H. currently serves on the scientific advisory board of Accure, Alector, and Annexon; board of directors of Neurona; and has previously consulted for BD, Moderna, and NGM Bio. Dr. Hauser also has received travel reimbursement and writing support from F. Hoffmann-La Roche and Novartis AG for anti-CD20 therapy-related meetings and presentations and is supported by grants from the NIH/NINDS (R35NS111644). R.G. H. has received fees for consultation from Roche/Genentech, Novartis, Neuron23, QIA Consulting, and research funding from Roche/Genentech and Atara. H.L. has received research support from the US Department of Defense and Octave Bioscience, Inc. A.P. is currently an employee of Moderna Therapeutics. F.Q. is an employee of Octave Bioscience, Inc. N.S. has received the Sylvia Lawry Physician Fellowship Award from the National MS Society. She has also received compensation for consulting from EMD Serono. H.W. has received research support from the Department of Defense, Genentech, Inc., National Institutes of Health, National Multiple Sclerosis Society, Novartis, and Sanofi Genzyme. He has received compensation for consulting from Genentech, Inc., IM Therapeutics, IMAB Biopharma, MedDay Pharmaceuticals, Tiziana Life Sciences, and vTv Therapeutics. S.E.B. is co-Founder of Mate Bioservices. H.Y., R.G., S.S., S.J.C., and A.S. declare no competing interests.
Figures


Update of
-
Inflammatory and neurodegenerative serum protein biomarkers increase sensitivity to detect disease activity in multiple sclerosis.medRxiv [Preprint]. 2023 Jul 3:2023.06.28.23291157. doi: 10.1101/2023.06.28.23291157. medRxiv. 2023. Update in: Nat Commun. 2024 May 20;15(1):4297. doi: 10.1038/s41467-024-48602-9. PMID: 37461671 Free PMC article. Updated. Preprint.

