Co-phylogeographic structure in a disease-causing parasite and its oyster host
- PMID: 38769826
- PMCID: PMC11474014
- DOI: 10.1017/S0031182024000611
Co-phylogeographic structure in a disease-causing parasite and its oyster host
Abstract
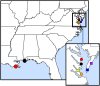
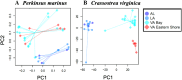
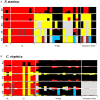
With the increasing affordability of next-generation sequencing technologies, genotype-by-sequencing has become a cost-effective tool for ecologists and conservation biologists to describe a species' evolutionary history. For host–parasite interactions, genotype-by-sequencing can allow the simultaneous examination of host and parasite genomes and can yield insight into co-evolutionary processes. The eastern oyster, Crassostrea virginica, is among the most important aquacultured species in the United States. Natural and farmed oyster populations can be heavily impacted by ‘dermo’ disease caused by an alveolate protist, Perkinsus marinus. Here, we used restricted site-associated DNA sequencing (RADseq) to simultaneously examine spatial population genetic structure of host and parasite. We analysed 393 single-nucleotide polymorphisms (SNPs) for P. marinus and 52,100 SNPs for C. virginica from 36 individual oysters from the Gulf of Mexico (GOM) and mid-Atlantic coastline. All analyses revealed statistically significant genetic differentiation between the GOM and mid-Atlantic coast populations for both C. virginica and P. marinus, and genetic divergence between Chesapeake Bay and the outer coast of Virginia for C. virginica, but not for P. marinus. A co-phylogenetic analysis confirmed significant coupled evolutionary change between host and parasite across large spatial scales. The strong genetic divergence between marine basins raises the possibility that oysters from either basin would not be well adapted to parasite genotypes and phenotypes from the other, which would argue for caution with regard to both oyster and parasite transfers between the Atlantic and GOM regions. More broadly, our results demonstrate the potential of RADseq to describe spatial patterns of genetic divergence consistent with coupled evolution.
Keywords: Crassostrea virginica; Perkinsus marinus; co-phylogeny; genetic divergence; genotype-by-sequencing.
Conflict of interest statement
None.
Figures


References
-
- Ansari MA, Pedergnana V, Ip LC, Magri A, Von Delft A, Bonsall D, Chaturvedi N, Bartha I, Smith D, Nicholson G, McVean G, Trebes A, Piazza P, Fellay J, Cooke G, Foster GR, Hudson E, McLauchlan J, Simmonds P, Bowden R, Klenerman P, Barnes E and Spencer CCA (2017) Genome-to-genome analysis highlights the effect of the human innate and adaptive immune systems on the hepatitis C virus. Nature Genetics 49, 666–673. - PMC - PubMed
-
- Archer FI, Adams PE, and Schneiders BB (2017) stratag: An r package for manipulating, summarizing and analysing population genetic data. Molecular Ecology Resources 17, 5–11. - PubMed
-
- Burford MO, Scarpa J, Cook BJ and Hare MP (2014) Local adaptation of a marine invertebrate with a high dispersal potential: evidence from a reciprocal transplant experiment of the eastern oyster Crassostrea virginica. Marine Ecology Progress Series 505, 161–175.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

