Bacterial cell differentiation enables population level survival strategies
- PMID: 38771034
- PMCID: PMC11237816
- DOI: 10.1128/mbio.00758-24
Bacterial cell differentiation enables population level survival strategies
Abstract
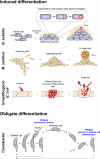
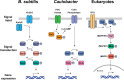
Clonal reproduction of unicellular organisms ensures the stable inheritance of genetic information. However, this means of reproduction lacks an intrinsic basis for genetic variation, other than spontaneous mutation and horizontal gene transfer. To make up for this lack of genetic variation, many unicellular organisms undergo the process of cell differentiation to achieve phenotypic heterogeneity within isogenic populations. Cell differentiation is either an inducible or obligate program. Induced cell differentiation can occur as a response to a stimulus, such as starvation or host cell invasion, or it can be a stochastic process. In contrast, obligate cell differentiation is hardwired into the organism's life cycle. Whether induced or obligate, bacterial cell differentiation requires the activation of a signal transduction pathway that initiates a global change in gene expression and ultimately results in a morphological change. While cell differentiation is considered a hallmark in the development of multicellular organisms, many unicellular bacteria utilize this process to implement survival strategies. In this review, we describe well-characterized cell differentiation programs to highlight three main survival strategies used by bacteria capable of differentiation: (i) environmental adaptation, (ii) division of labor, and (iii) bet-hedging.
Keywords: Bacillus subtilis; Caulobacter crescentus; Myxococcus xanthus; bet-hedging; cell differentiation; division of labor; signal transduction.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Bet hedging or not? A guide to proper classification of microbial survival strategies.Bioessays. 2011 Mar;33(3):215-23. doi: 10.1002/bies.201000127. Epub 2011 Jan 21. Bioessays. 2011. PMID: 21254151
-
Phenotypic Heterogeneity in Bacterial Quorum Sensing Systems.J Mol Biol. 2019 Nov 22;431(23):4530-4546. doi: 10.1016/j.jmb.2019.04.036. Epub 2019 Apr 30. J Mol Biol. 2019. PMID: 31051177 Review.
-
The deprivation syndrome is the driving force of phylogeny, ontogeny and oncogeny.Rev Neurosci. 2001;12(3):217-87. doi: 10.1515/revneuro.2001.12.3.217. Rev Neurosci. 2001. PMID: 11560369 Review.
-
Coordinated Cell Death in Isogenic Bacterial Populations: Sacrificing Some for the Benefit of Many?J Mol Biol. 2019 Nov 22;431(23):4656-4669. doi: 10.1016/j.jmb.2019.04.024. Epub 2019 Apr 25. J Mol Biol. 2019. PMID: 31029705 Review.
-
Bacterial differentiation.Science. 1971 Sep 3;173(4000):884-92. doi: 10.1126/science.173.4000.884. Science. 1971. PMID: 5572165
Cited by
-
Chemical Conversations.Molecules. 2025 Jan 21;30(3):431. doi: 10.3390/molecules30030431. Molecules. 2025. PMID: 39942538 Free PMC article. Review.
-
The development of Burkholderia bacteria as heterologous hosts.Nat Prod Rep. 2025 Jul 28. doi: 10.1039/d5np00024f. Online ahead of print. Nat Prod Rep. 2025. PMID: 40718961 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
