ADAM17 variant causes hair loss via ubiquitin ligase TRIM47-mediated degradation
- PMID: 38771644
- PMCID: PMC11383180
- DOI: 10.1172/jci.insight.177588
ADAM17 variant causes hair loss via ubiquitin ligase TRIM47-mediated degradation
Abstract
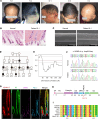
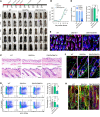
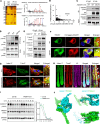
Hypotrichosis is a genetic disorder characterized by a diffuse and progressive loss of scalp and/or body hair. Nonetheless, the causative genes for several affected individuals remain elusive, and the underlying mechanisms have yet to be fully elucidated. Here, we discovered a dominant variant in a disintegrin and a metalloproteinase domain 17 (ADAM17) gene caused hypotrichosis with woolly hair. Adam17 (p.D647N) knockin mice mimicked the hair abnormality in patients. ADAM17 (p.D647N) mutation led to hair follicle stem cell (HFSC) exhaustion and caused abnormal hair follicles, ultimately resulting in alopecia. Mechanistic studies revealed that ADAM17 binds directly to E3 ubiquitin ligase tripartite motif-containing protein 47 (TRIM47). ADAM17 variant enhanced the association between ADAM17 and TRIM47, leading to an increase in ubiquitination and subsequent degradation of ADAM17 protein. Furthermore, reduced ADAM17 protein expression affected the Notch signaling pathway, impairing the activation, proliferation, and differentiation of HFSCs during hair follicle regeneration. Overexpression of Notch intracellular domain rescued the reduced proliferation ability caused by Adam17 variant in primary fibroblast cells.
Keywords: Dermatology; Genetic diseases; Genetic variation; Genetics; Ubiquitin-proteosome system.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

