Multidrug-resistance and extended-spectrum beta-lactamase-producing lactose-fermenting enterobacteriaceae in the human-dairy interface in northwest Ethiopia
- PMID: 38771780
- PMCID: PMC11108214
- DOI: 10.1371/journal.pone.0303872
Multidrug-resistance and extended-spectrum beta-lactamase-producing lactose-fermenting enterobacteriaceae in the human-dairy interface in northwest Ethiopia
Abstract
Background: Antimicrobial resistance (AMR) is among the top public health concerns in the globe. Estimating the prevalence of multidrug resistance (MDR), MDR index (MDR-I) and extended-spectrum beta-lactamase (ESBL)-producing lactose fermenting Enterobacteriaceae (LFE) is important in designing strategies to combat AMR. Thus, this study was designed to determine the status of MDR, MDR-I and ESBL-producing LFE isolated from the human-dairy interface in the northwestern part of Ethiopia, where such information is lacking.
Methodology: A cross-sectional study was conducted from June 2022 to August 2023 by analyzing 362 samples consisting of raw pooled milk (58), milk container swabs (58), milker's hand swabs (58), farm sewage (57), milker's stool (47), and cow's feces (84). The samples were analyzed using standard bacteriological methods. The antimicrobial susceptibility patterns and ESBL production ability of the LFE isolates were screened using the Kirby-Bauer disk diffusion method, and candidate isolates passing the screening criteria were phenotypically confirmed by using cefotaxime (30 μg) and cefotaxime /clavulanic acid (30 μg/10 μg) combined-disk diffusion test. The isolates were further characterized genotypically using multiplex polymerase chain reaction targeting the three ESBL-encoding- genes namely blaTEM, blaSHV, and blaCTX-M.
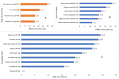
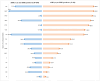
Results: A total of 375 bacterial isolates were identified and the proportion of MDR and ESBL-producing bacterial isolates were 70.7 and 21.3%, respectively. The MDR-I varied from 0.0 to 0.81 with an average of 0.30. The ESBL production was detected in all sample types. Genotypically, the majority of the isolates (97.5%), which were positive on the phenotypic test, were carrying one or more of the three genes.
Conclusion: A high proportion of the bacterial isolates were MDR; had high MDR-I and were positive for ESBL production. The findings provide evidence that the human-dairy interface is one of the important reservoirs of AMR traits. Therefore, the implementation of AMR mitigation strategies is highly needed in the area.
Copyright: This is an open access article, free of all copyright, and may be freely reproduced, distributed, transmitted, modified, built upon, or otherwise used by anyone for any lawful purpose. The work is made available under the Creative Commons CC0 public domain dedication.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- FAO/WHO. Shiga toxin-producing Escherichia coli (STEC) and food: attribution, characterization, and monitoring: Risk, Microbiological Assessment Series, Assessment, https://apps.who.int/iris/bitstream/handle/10665/272871/9789241514279-en.... 2018.
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

