Analysis of common differential gene expression in synovial cells of osteoarthritis and rheumatoid arthritis
- PMID: 38771826
- PMCID: PMC11108184
- DOI: 10.1371/journal.pone.0303506
Analysis of common differential gene expression in synovial cells of osteoarthritis and rheumatoid arthritis
Abstract
Objective: To elucidate potential molecular mechanisms differentiating osteoarthritis (OA) and rheumatoid arthritis (RA) through a bioinformatics analysis of differentially expressed genes (DEGs) in patient synovial cells, aiming to provide new insights for clinical treatment strategies.
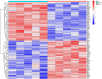
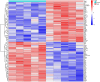
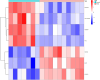
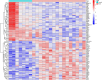
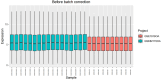
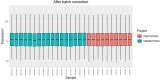
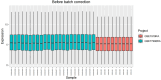
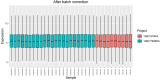
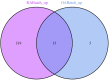
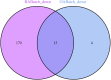
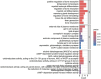
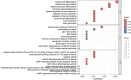
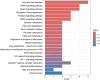
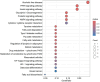
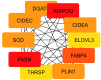
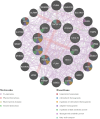
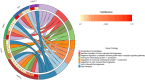
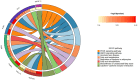
Materials and methods: Gene expression datasets GSE1919, GSE82107, and GSE77298 were downloaded from the Gene Expression Omnibus (GEO) database to serve as the training groups, with GSE55235 being used as the validation dataset. The OA and RA data from the GSE1919 dataset were merged with the standardized data from GSE82107 and GSE77298, followed by batch effect removal to obtain the merged datasets of differential expressed genes (DEGs) for OA and RA. Intersection analysis was conducted on the DEGs between the two conditions to identify commonly upregulated and downregulated DEGs. Enrichment analysis was then performed on these common co-expressed DEGs, and a protein-protein interaction (PPI) network was constructed to identify hub genes. These hub genes were further analyzed using the GENEMANIA online platform and subjected to enrichment analysis. Subsequent validation analysis was conducted using the GSE55235 dataset.
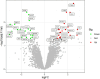
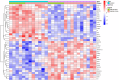
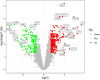
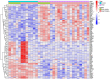
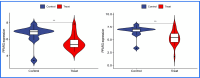
Results: The analysis of differentially expressed genes in the synovial cells from patients with Osteoarthritis (OA) and Rheumatoid Arthritis (RA), compared to a control group (individuals without OA or RA), revealed significant changes in gene expression patterns. Specifically, the genes APOD, FASN, and SCD were observed to have lower expression levels in the synovial cells of both OA and RA patients, indicating downregulation within the pathological context of these diseases. In contrast, the SDC1 gene was found to be upregulated, displaying higher expression levels in the synovial cells of OA and RA patients compared to normal controls.Additionally, a noteworthy observation was the downregulation of the transcription factor PPARG in the synovial cells of patients with OA and RA. The decrease in expression levels of PPARG further validates the alteration in lipid metabolism and inflammatory processes associated with the pathogenesis of OA and RA. These findings underscore the significance of these genes and the transcription factor not only as biomarkers for differential diagnosis between OA and RA but also as potential targets for therapeutic interventions aimed at modulating their expression to counteract disease progression.
Conclusion: The outcomes of this investigation reveal the existence of potentially shared molecular mechanisms within Osteoarthritis (OA) and Rheumatoid Arthritis (RA). The identification of APOD, FASN, SDC1, TNFSF11 as key target genes, along with their downstream transcription factor PPARG, highlights common potential factors implicated in both diseases. A deeper examination and exploration of these findings could pave the way for new candidate targets and directions in therapeutic research aimed at treating both OA and RA. This study underscores the significance of leveraging bioinformatics approaches to unravel complex disease mechanisms, offering a promising avenue for the development of more effective and targeted treatments.
Copyright: © 2024 Liao et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Bliddal H. [Definition, pathology and pathogenesis of osteoarthritis]. Ugeskr Laeger. 2020. Oct 12;182(42):V06200477. Danish. . - PubMed
-
- Roberts NLS, Mountjoy-Venning WC, Anjomshoa M, Banoub JAM, Yasin YJ. GBD 2017 Disease and Injury Incidence and Prevalence Collaborators. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study (vol 392, pg 1789, 2018) Lancet. 2019;393:E44–E. - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

