Type VII secretion system extracellular protein B targets STING to evade host anti- Staphylococcus aureus immunity
- PMID: 38771879
- PMCID: PMC11145284
- DOI: 10.1073/pnas.2402764121
Type VII secretion system extracellular protein B targets STING to evade host anti- Staphylococcus aureus immunity
Abstract
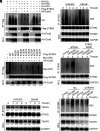
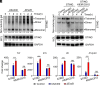
Staphylococcus aureus (S. aureus) can evade antibiotics and host immune defenses by persisting within infected cells. Here, we demonstrate that in infected host cells, S. aureus type VII secretion system (T7SS) extracellular protein B (EsxB) interacts with the stimulator of interferon genes (STING) protein and suppresses the inflammatory defense mechanism of macrophages during early infection. The binding of EsxB with STING disrupts the K48-linked ubiquitination of EsxB at lysine 33, thereby preventing EsxB degradation. Furthermore, EsxB-STING binding appears to interrupt the interaction of 2 vital regulatory proteins with STING: aspartate-histidine-histidine-cysteine domain-containing protein 3 (DHHC3) and TNF receptor-associated factor 6. This persistent dual suppression of STING interactions deregulates intracellular proinflammatory pathways in macrophages, inhibiting STING's palmitoylation at cysteine 91 and its K63-linked ubiquitination at lysine 83. These findings uncover an immune-evasion mechanism by S. aureus T7SS during intracellular macrophage infection, which has implications for developing effective immunomodulators to combat S. aureus infections.
Keywords: EsxB; STING; Staphylococcus aureus; inflammation; macrophages.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures
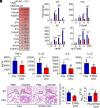
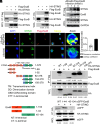
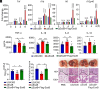
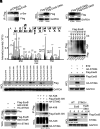

References
MeSH terms
Substances
Grants and funding
- 2021YFC2301405/MOST | National Key Research and Development Program of China (NKPs)
- 82201942/MOST | National Natural Science Foundation of China (NSFC)
- 32070941/MOST | National Natural Science Foundation of China (NSFC)
- 32270989/MOST | National Natural Science Foundation of China (NSFC)
- CQYC202003220/technology innovation and application development project of Chongqing
LinkOut - more resources
Full Text Sources
Medical
Research Materials

