Single Cell Analysis of Human Colonoids Exposed to Uranium-Bearing Dust
- PMID: 38771937
- PMCID: PMC11108582
- DOI: 10.1289/EHP13855
Single Cell Analysis of Human Colonoids Exposed to Uranium-Bearing Dust
Abstract
Background: Uranium exposure remains an important environmental legacy and physiological health concern, with hundreds of abandoned uranium mines located in the Southwestern United States largely impacting underserved indigenous communities. The negative effects of heavy metals on barrier permeability and inhibition of intestinal epithelial healing have been described; however, transcriptomic changes within the intestinal epithelial cells and impacts on lineage differentiation are largely unknown.
Objectives: Herein, we sought to determine the molecular and cellular changes that occur in the colon in response to uranium bearing dust (UBD) exposure.
Methods: Human colonoids from three biologically distinct donors were acutely exposed to UBD then digested for single cell RNA sequencing to define the molecular changes that occur to specific identities of colonic epithelial cells. Validation in colonoids was assessed using morphological and imaging techniques.
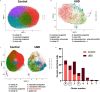
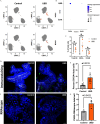
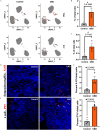
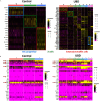
Results: Human colonoids acutely exposed to UBD exhibited disrupted proliferation and hyperplastic differentiation of the secretory lineage cell, enteroendocrine cells (EEC). Single-cell RNA sequencing also showed more EEC subtypes present in UBD-exposed colonoids.
Discussion: These findings highlight the significance of crypt-based proliferative cells and secretory cell differentiation using human colonoids to model major colonic responses to uranium-bearing particulate dust exposure. https://doi.org/10.1289/EHP13855.
Figures



References
-
- Lin Y, Hoover J, Beene D, Erdei E, Liu Z. 2020. Environmental risk mapping of potential abandoned uranium mine contamination on the Navajo nation, USA, using a GIS-based multi-criteria decision analysis approach. Environ Sci Pollut Res Int 27(24):30542–30557, PMID: 32468361, 10.1007/s11356-020-09257-3. - DOI - PMC - PubMed
-
- EPA (US Environmental Protection Agency). 2006. Basic Information about Radionuclides in Drinking Water. Washington, DC: EPA.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
