Anti-EGFR Antibody-Drug Conjugate Carrying an Inhibitor Targeting CDK Restricts Triple-Negative Breast Cancer Growth
- PMID: 38772416
- PMCID: PMC11292198
- DOI: 10.1158/1078-0432.CCR-23-3110
Anti-EGFR Antibody-Drug Conjugate Carrying an Inhibitor Targeting CDK Restricts Triple-Negative Breast Cancer Growth
Abstract
Purpose: Anti-EGFR antibodies show limited response in breast cancer, partly due to activation of compensatory pathways. Furthermore, despite the clinical success of cyclin-dependent kinase (CDK) 4/6 inhibitors in hormone receptor-positive tumors, aggressive triple-negative breast cancers (TNBC) are largely resistant due to CDK2/cyclin E expression, whereas free CDK2 inhibitors display normal tissue toxicity, limiting their therapeutic application. A cetuximab-based antibody drug conjugate (ADC) carrying a CDK inhibitor selected based on oncogene dysregulation, alongside patient subgroup stratification, may provide EGFR-targeted delivery.
Experimental design: Expressions of G1/S-phase cell cycle regulators were evaluated alongside EGFR in breast cancer. We conjugated cetuximab with CDK inhibitor SNS-032, for specific delivery to EGFR-expressing cells. We assessed ADC internalization and its antitumor functions in vitro and in orthotopically grown basal-like/TNBC xenografts.
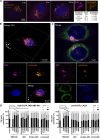
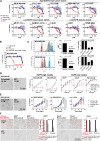
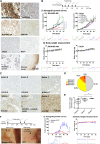
Results: Transcriptomic (6,173 primary, 27 baseline, and matched post-chemotherapy residual tumors), single-cell RNA sequencing (150,290 cells, 27 treatment-naïve tumors), and spatial transcriptomic (43 tumor sections, 22 TNBCs) analyses confirmed expression of CDK2 and its cyclin partners in basal-like/TNBCs, associated with EGFR. Spatiotemporal live-cell imaging and super-resolution confocal microscopy demonstrated ADC colocalization with late lysosomal clusters. The ADC inhibited cell cycle progression, induced cytotoxicity against high EGFR-expressing tumor cells, and bystander killing of neighboring EGFR-low tumor cells, but minimal effects on immune cells. Despite carrying a small molar fraction (1.65%) of the SNS-032 inhibitor, the ADC restricted EGFR-expressing spheroid and cell line/patient-derived xenograft tumor growth.
Conclusions: Exploiting EGFR overexpression, and dysregulated cell cycle in aggressive and treatment-refractory tumors, a cetuximab-CDK inhibitor ADC may provide selective and efficacious delivery of cell cycle-targeted agents to basal-like/TNBCs, including chemotherapy-resistant residual disease.
©2024 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
A. Cheung reports grants from Breast Cancer Now during the conduct of the study; in addition, A. Cheung has a patent for antibody technology pending. A.M. Chenoweth reports grants from Breast Cancer Now during the conduct of the study; in addition, A.M. Chenoweth has a patent for antibody technologies pending. P. Romero-Clavijo reports grants from CRUK City of London Cancer Centre during the conduct of the study. K.M. Rahman reports grants and other support from Pheon Therapeutics outside the submitted work; in addition, K.M. Rahman has a patent for US10399970B2 issued, US10975072B2 issued, US10975074B2 issued, US9999625B2 issued, and US9376440B2 issued. C.J. Lord reports grants and personal fees from AstraZeneca, Merck KGaA, Artios, and Neophore, as well as personal fees from ForEx, Syncona, Sun Pharma, 3rd Rock, Ono Pharma, Abingworth, Dark Blue Therapeutics, Pontifax, Astex, GlaxoSmithKline, Dawn Bioventures, Blacksmith Medicines, Gerson Lehrman Group, and Vertex; personal fees and other support from Tango Therapeutics and Tesselate; and other support from Ovibio and Hysplex outside the submitted work. C.J. Lord is also a named inventor on patents describing the use of DNA repair inhibitors and stands to gain from their development and use as part of the ICR “Rewards to Inventors” scheme and also reports benefits from this scheme associated with patents for PARP inhibitors paid into C.J. Lord’s personal account and research accounts at the Institute of Cancer Research. A.N.J. Tutt reports grants from Breast Cancer Now during the conduct of the study; other support from The Institute of Cancer Research, Tango, and Pfizer; grants, personal fees, and other support from AstraZeneca; and nonfinancial support and other support from Guardant Health outside the submitted work. S.N. Karagiannis reports grants from Breast Cancer Now, The Biotechnology and Biological Sciences Research Council, Cancer Research UK, the Medical Research Council, National Institute for Health Research (NIHR), and Worldwide Cancer Research during the conduct of the study, as well as grants from Epsilogen Ltd. outside the submitted work; in addition, S.N. Karagiannis declares patents for antibody technologies granted and pending and is founder and shareholder of Epsilogen Ltd. No disclosures were reported by the other authors.
Figures



References
-
- Emens LA, Adams S, Barrios CH, Dieras V, Iwata H, Loi S, et al. . First-line atezolizumab plus nab-paclitaxel for unresectable, locally advanced, or metastatic triple-negative breast cancer: IMpassion130 final overall survival analysis. Ann Oncol 2021;32:983–93. - PubMed
-
- Bardia A, Hurvitz SA, Tolaney SM, Loirat D, Punie K, Oliveira M, et al. . Sacituzumab govitecan in metastatic triple-negative breast cancer. N Engl J Med 2021;384:1529–41. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

