Determinants of gastric cancer immune escape identified from non-coding immune-landscape quantitative trait loci
- PMID: 38773080
- PMCID: PMC11109163
- DOI: 10.1038/s41467-024-48436-5
Determinants of gastric cancer immune escape identified from non-coding immune-landscape quantitative trait loci
Abstract
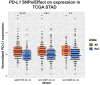
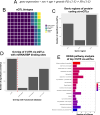
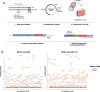
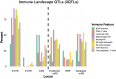
The landscape of non-coding mutations in cancer progression and immune evasion is largely unexplored. Here, we identify transcrptome-wide somatic and germline 3' untranslated region (3'-UTR) variants from 375 gastric cancer patients from The Cancer Genome Atlas. By performing gene expression quantitative trait loci (eQTL) and immune landscape QTL (ilQTL) analysis, we discover 3'-UTR variants with cis effects on expression and immune landscape phenotypes, such as immune cell infiltration and T cell receptor diversity. Using a massively parallel reporter assay, we distinguish between causal and correlative effects of 3'-UTR eQTLs in immune-related genes. Our approach identifies numerous 3'-UTR eQTLs and ilQTLs, providing a unique resource for the identification of immunotherapeutic targets and biomarkers. A prioritized ilQTL variant signature predicts response to immunotherapy better than standard-of-care PD-L1 expression in independent patient cohorts, showcasing the untapped potential of non-coding mutations in cancer.
© 2024. The Author(s).
Conflict of interest statement
I.S.V. consults for Guidepoint Global, Cowen, Mosaic, and NextRNA. Beth Israel Deaconess Medical Center has filed a patent application based on this work for “Methods and compositions related to non-coding variants for the prediction of response to cancer immunotherapy” under 63/378,392, where I.S.V., F.J.S., C.M., and E.K. are named as co-inventors. The remaining authors declare no competing interests.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

