Astrocytic ALKBH5 in stress response contributes to depressive-like behaviors in mice
- PMID: 38773146
- PMCID: PMC11109195
- DOI: 10.1038/s41467-024-48730-2
Astrocytic ALKBH5 in stress response contributes to depressive-like behaviors in mice
Abstract
Epigenetic mechanisms bridge genetic and environmental factors that contribute to the pathogenesis of major depression disorder (MDD). However, the cellular specificity and sensitivity of environmental stress on brain epitranscriptomics and its impact on depression remain unclear. Here, we found that ALKBH5, an RNA demethylase of N6-methyladenosine (m6A), was increased in MDD patients' blood and depression models. ALKBH5 in astrocytes was more sensitive to stress than that in neurons and endothelial cells. Selective deletion of ALKBH5 in astrocytes, but not in neurons and endothelial cells, produced antidepressant-like behaviors. Astrocytic ALKBH5 in the mPFC regulated depression-related behaviors bidirectionally. Meanwhile, ALKBH5 modulated glutamate transporter-1 (GLT-1) m6A modification and increased the expression of GLT-1 in astrocytes. ALKBH5 astrocyte-specific knockout preserved stress-induced disruption of glutamatergic synaptic transmission, neuronal atrophy and defective Ca2+ activity. Moreover, enhanced m6A modification with S-adenosylmethionine (SAMe) produced antidepressant-like effects. Our findings indicate that astrocytic epitranscriptomics contribute to depressive-like behaviors and that astrocytic ALKBH5 may be a therapeutic target for depression.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

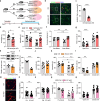




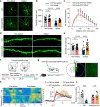
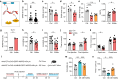
References
-
- World, H.O. Depression and Other Common Mental Disorders: Global Health Estimates (World Health Organization, 2017).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

