Svetlana a supervised segmentation classifier for Napari
- PMID: 38773203
- PMCID: PMC11109332
- DOI: 10.1038/s41598-024-60916-8
Svetlana a supervised segmentation classifier for Napari
Abstract
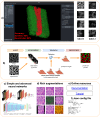
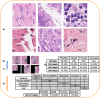
We present Svetlana (SuperVised sEgmenTation cLAssifier for NapAri), an open-source Napari plugin dedicated to the manual or automatic classification of segmentation results. A few recent software tools have made it possible to automatically segment complex 2D and 3D objects such as cells in biology with unrivaled performance. However, the subsequent analysis of the results is oftentimes inaccessible to non-specialists. The Svetlana plugin aims at going one step further, by allowing end-users to label the segmented objects and to pick, train and run arbitrary neural network classifiers. The resulting network can then be used for the quantitative analysis of biophysical phenoma. We showcase its performance through challenging problems in 2D and 3D and provide a comprehensive discussion on its strengths and limits.
Keywords: Biomedical imaging; Classification; Convolutional neural networks; Efficient AI; Image analysis; Microscopy; Segmentation; Software.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

