Unraveling resistance mechanisms in anti-CD19 chimeric antigen receptor-T therapy for B-ALL: a novel in vitro model and insights into target antigen dynamics
- PMID: 38773607
- PMCID: PMC11110321
- DOI: 10.1186/s12967-024-05254-z
Unraveling resistance mechanisms in anti-CD19 chimeric antigen receptor-T therapy for B-ALL: a novel in vitro model and insights into target antigen dynamics
Abstract
Background: Cellular immunotherapy, represented by the chimeric antigen receptor T cell (CAR-T), has exhibited high response rates, durable remission, and safety in vitro and in clinical trials. Unfortunately, anti-CD19 CAR-T (CART-19) treatment alone is prone to relapse and has a particularly poor prognosis in relapsed/refractory (r/r) B-ALL patients. To date, addressing or reducing relapse remains one of the research priorities to achieve broad clinical application.
Methods: We manufactured second generation CART-19 cells and validated their efficacy and safety in vitro and in vivo. Through co-culture of Nalm-6 cells with short-term cultured CART-19 cells, CD19-negative Nalm-6 cells were detected by flow cytometry, and further investigation of the relapsed cells and their resistance mechanisms was evaluated in vitro.
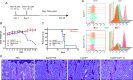
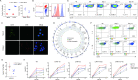
Results: In this study, we demonstrated that CART-19 cells had enhanced and specific antileukemic activities, and the survival of B-ALL mouse models after CART-19 treatment was significantly prolonged. We then shortened the culture time and applied the serum-free culture to expand CAR-T cells, followed by co-culturing CART-19 cells with Nalm-6 cells. Surprisingly, we observed the proliferation of CD19-negative Nalm-6 cells around 28 days. Identification of potential resistance mechanisms showed that the relapsed cells express truncated CD19 proteins with decreased levels and, more importantly, CAR expression was detected on the relapsed cell surface, which may ultimately keep them antigen-negative. Furthermore, it was validated that CART-22 and tandem CART-22/19 cells could effectively kill the relapsed cells, but neither could completely eradicate them.
Conclusions: We successfully generated CART-19 cells and obtained a CD19-negative refractory relapsed B-ALL cell line, providing new insights into the underlying mechanisms of resistance and a new in vitro model for the treatment of r/r B-ALL patients with low antigen density.
Keywords: Antigen negative relapse; B-ALL; CAR-T cell therapy; CD19; Resistant mechanisms.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

