A SAW-Based Programmable Controlled RNA Detecting Device: Rapid In Situ Cytolysis-RNA Capture-RNA Release-PCR in One Mini Chamber
- PMID: 38773709
- PMCID: PMC11304306
- DOI: 10.1002/advs.202309744
A SAW-Based Programmable Controlled RNA Detecting Device: Rapid In Situ Cytolysis-RNA Capture-RNA Release-PCR in One Mini Chamber
Abstract
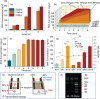
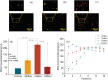
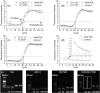
Viral RNA detection is crucial in preventing and treating early infectious diseases. Traditional methods of RNA detection require a large amount of equipment and technical personnel. In this study, proposed a programmable controlled surface acoustic wave (SAW)-based RNA detecting device has been proposed. The proposed device can perform the entire viral RNA detection process, including cell lysis by cell-microparticle collision through SAW-induced liquid whirling, RNA capture by SAW-suspended magnetic beads, RNA elution through SAW-induced high streaming force, and PCR thermal cycling through SAW-generated heat. The device has completed all RNA detection steps in one mini chamber, requiring only 489 µl reagents for RNA extraction, much smaller than the amount used in manual RNA extraction (2065 µl). The experimental results have shown that PCR results from the device are comparable to those achieved via commercial qPCR instrumental detection. This work has demonstrated the potential of SAW-based lab-on-a-chip devices for point-of-care testing and provided a novel approach for rapidly detecting infectious diseases.
Keywords: RNA detection; microfluidic; surface acoustic wave.
© 2024 The Authors. Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







Similar articles
-
Rapid acoustofluidic mixing by ultrasonic surface acoustic wave-induced acoustic streaming flow.Ultrason Sonochem. 2023 Oct;99:106575. doi: 10.1016/j.ultsonch.2023.106575. Epub 2023 Sep 4. Ultrason Sonochem. 2023. PMID: 37683414 Free PMC article.
-
[Standard technical specifications for methacholine chloride (Methacholine) bronchial challenge test (2023)].Zhonghua Jie He He Hu Xi Za Zhi. 2024 Feb 12;47(2):101-119. doi: 10.3760/cma.j.cn112147-20231019-00247. Zhonghua Jie He He Hu Xi Za Zhi. 2024. PMID: 38309959 Chinese.
-
Planar chip device for PCR and hybridization with surface acoustic wave pump.Lab Chip. 2005 Mar;5(3):308-17. doi: 10.1039/b412712a. Epub 2004 Dec 16. Lab Chip. 2005. PMID: 15726207
-
Surface acoustic wave (SAW) techniques in tissue engineering.Cell Tissue Res. 2021 Nov;386(2):215-226. doi: 10.1007/s00441-020-03397-1. Epub 2021 Aug 14. Cell Tissue Res. 2021. PMID: 34390407 Review.
-
Implementation of guiding layers of surface acoustic wave devices: A review.Biosens Bioelectron. 2018 Jan 15;99:500-512. doi: 10.1016/j.bios.2017.07.060. Epub 2017 Jul 29. Biosens Bioelectron. 2018. PMID: 28823975 Review.
Cited by
-
Acoustofluidics: Technology Advances and Applications from 2022 to 2024.Anal Chem. 2025 Apr 8;97(13):6847-6870. doi: 10.1021/acs.analchem.4c06803. Epub 2025 Mar 25. Anal Chem. 2025. PMID: 40133046 Free PMC article. Review. No abstract available.
References
-
- Oeschger T. M., McCloskey D. S., Buchmann R. M., Choubal A. M., Boza J. M., Mehta S., Erickson D., Acc. Chem. Res. 2021, 54, 3656. - PubMed
-
- Trinh T. N. D., Lee N. Y., Chemosensors 2022, 10, 123.
-
- Wu J., He Z., Chen Q., Lin J.‐M., TrAC, Trends Anal. Chem. 2016, 80, 213.
MeSH terms
Substances
Grants and funding
- 2023YFB3210400/National Key Research and Development Plan of China
- 2022B1111010003/Key-Area Research and Development Program of Guangdong Province
- 62273329/National Natural Science Foundation of China
- 62050410349/National Natural Science Foundation of China
- JCYJ20210324115601004/Key Fundamental Research Program of Shenzhen
LinkOut - more resources
Full Text Sources
Miscellaneous
