Spatial Transcriptomics: A Powerful Tool in Disease Understanding and Drug Discovery
- PMID: 38773973
- PMCID: PMC11103497
- DOI: 10.7150/thno.95908
Spatial Transcriptomics: A Powerful Tool in Disease Understanding and Drug Discovery
Abstract
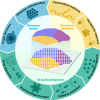
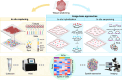
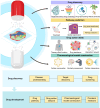
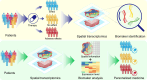
Recent advancements in modern science have provided robust tools for drug discovery. The rapid development of transcriptome sequencing technologies has given rise to single-cell transcriptomics and single-nucleus transcriptomics, increasing the accuracy of sequencing and accelerating the drug discovery process. With the evolution of single-cell transcriptomics, spatial transcriptomics (ST) technology has emerged as a derivative approach. Spatial transcriptomics has emerged as a hot topic in the field of omics research in recent years; it not only provides information on gene expression levels but also offers spatial information on gene expression. This technology has shown tremendous potential in research on disease understanding and drug discovery. In this article, we introduce the analytical strategies of spatial transcriptomics and review its applications in novel target discovery and drug mechanism unravelling. Moreover, we discuss the current challenges and issues in this research field that need to be addressed. In conclusion, spatial transcriptomics offers a new perspective for drug discovery.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures


References
-
- Ståhl PL, Salmén F, Vickovic S. et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016;353:78–82. - PubMed
-
- Strell C, Hilscher MM, Laxman N. et al. Placing RNA in context and space - methods for spatially resolved transcriptomics. FEBS J. 2019;286:1468–81. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

