Small extracellular vesicles (sEVs)-based gene delivery platform for cell-specific CRISPR/Cas9 genome editing
- PMID: 38773978
- PMCID: PMC11103490
- DOI: 10.7150/thno.92133
Small extracellular vesicles (sEVs)-based gene delivery platform for cell-specific CRISPR/Cas9 genome editing
Abstract
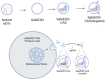
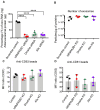
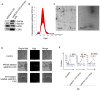
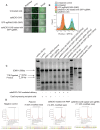
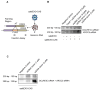
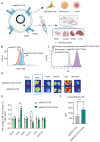
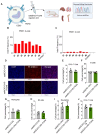
Small extracellular vesicles (sEVs) are naturally occurring vesicles that have the potential to be manipulated to become promising drug delivery vehicles for on-demand in vitro and in vivo gene editing. Here, we developed the modular safeEXO platform, a prototype sEV delivery vehicle that is mostly devoid of endogenous RNA and can efficaciously deliver RNA and ribonucleoprotein (RNP) complexes to their intended intracellular targets manifested by downstream biologic activity. We also successfully engineered producer cells to produce safeEXO vehicles that contain endogenous Cas9 (safeEXO-CAS) to effectively deliver efficient ribonucleoprotein (RNP)-mediated CRISPR genome editing machinery to organs or diseased cells in vitro and in vivo. We confirmed that safeEXO-CAS sEVs could co-deliver ssDNA, sgRNA and siRNA, and efficaciously mediate gene insertion in a dose-dependent manner. We demonstrated the potential to target safeEXO-CAS sEVs by engineering sEVs to express a tissue-specific moiety, integrin alpha-6 (safeEXO-CAS-ITGA6), which increased their uptake to lung epithelial cells in vitro and in vivo. We tested the ability of safeEXO-CAS-ITGA6 loaded with EMX1 sgRNAs to induce lung-targeted editing in mice, which demonstrated significant gene editing in the lungs with no signs of morbidity or detectable changes in immune cell populations. Our results demonstrate that our modular safeEXO platform represents a targetable, safe, and efficacious vehicle to deliver nucleic acid-based therapeutics that successfully reach their intracellular targets. Furthermore, safeEXO producer cells can be genetically manipulated to produce safeEXO vehicles containing CRISPR machinery for more efficient RNP-mediated genome editing. This platform has the potential to improve current therapies and increase the landscape of treatment for various human diseases using RNAi and CRISPR approaches.
Keywords: CRISPR; Cas; Delivery; Genome Editing; sEVs.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

