Identification of diverse RNA viruses in Obscuromonas flagellates (Euglenozoa: Trypanosomatidae: Blastocrithidiinae)
- PMID: 38774311
- PMCID: PMC11108086
- DOI: 10.1093/ve/veae037
Identification of diverse RNA viruses in Obscuromonas flagellates (Euglenozoa: Trypanosomatidae: Blastocrithidiinae)
Abstract
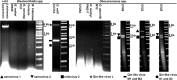
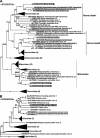
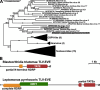
Trypanosomatids (Euglenozoa) are a diverse group of unicellular flagellates predominately infecting insects (monoxenous species) or circulating between insects and vertebrates or plants (dixenous species). Monoxenous trypanosomatids harbor a wide range of RNA viruses belonging to the families Narnaviridae, Totiviridae, Qinviridae, Leishbuviridae, and a putative group of tombus-like viruses. Here, we focus on the subfamily Blastocrithidiinae, a previously unexplored divergent group of monoxenous trypanosomatids comprising two related genera: Obscuromonas and Blastocrithidia. Members of the genus Blastocrithidia employ a unique genetic code, in which all three stop codons are repurposed to encode amino acids, with TAA also used to terminate translation. Obscuromonas isolates studied here bear viruses of three families: Narnaviridae, Qinviridae, and Mitoviridae. The latter viral group is documented in trypanosomatid flagellates for the first time. While other known mitoviruses replicate in the mitochondria, those of trypanosomatids appear to reside in the cytoplasm. Although no RNA viruses were detected in Blastocrithidia spp., we identified an endogenous viral element in the genome of B. triatomae indicating its past encounter(s) with tombus-like viruses.
Keywords: Blastocrithidia; Mitoviridae; Narnaviridae; Obscuromonas; Qin-like virus; dsRNA viruses.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures
Similar articles
-
Comparative genomic analysis of trypanosomatid protists illuminates an extensive change in the nuclear genetic code.mBio. 2025 Jun 11;16(6):e0088525. doi: 10.1128/mbio.00885-25. Epub 2025 Apr 28. mBio. 2025. PMID: 40293238 Free PMC article.
-
Diversity of RNA viruses in the cosmopolitan monoxenous trypanosomatid Leptomonas pyrrhocoris.BMC Biol. 2023 Sep 12;21(1):191. doi: 10.1186/s12915-023-01687-y. BMC Biol. 2023. PMID: 37697369 Free PMC article.
-
Characterization of a new cosmopolitan genus of trypanosomatid parasites, Obscuromonas gen. nov. (Blastocrithidiinae subfam. nov.).Eur J Protistol. 2021 Jun;79:125778. doi: 10.1016/j.ejop.2021.125778. Epub 2021 Feb 2. Eur J Protistol. 2021. PMID: 33706204
-
The evolution of trypanosomatid taxonomy.Parasit Vectors. 2017 Jun 8;10(1):287. doi: 10.1186/s13071-017-2204-7. Parasit Vectors. 2017. PMID: 28595622 Free PMC article. Review.
-
A Stroll Through the History of Monoxenous Trypanosomatids Infection in Vertebrate Hosts.Front Cell Infect Microbiol. 2022 Feb 15;12:804707. doi: 10.3389/fcimb.2022.804707. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35242719 Free PMC article. Review.
Cited by
-
A novel strain of Leishmania braziliensis harbors not a toti- but a bunyavirus.PLoS Negl Trop Dis. 2024 Dec 27;18(12):e0012767. doi: 10.1371/journal.pntd.0012767. eCollection 2024 Dec. PLoS Negl Trop Dis. 2024. PMID: 39729426 Free PMC article.
-
Comparative genomic analysis of trypanosomatid protists illuminates an extensive change in the nuclear genetic code.mBio. 2025 Jun 11;16(6):e0088525. doi: 10.1128/mbio.00885-25. Epub 2025 Apr 28. mBio. 2025. PMID: 40293238 Free PMC article.
-
Converting Blastocrithidia Nonstop, a Trypanosomatid With Non-Canonical Genetic Code, Into a Genetically-Tractable Model.Mol Microbiol. 2025 Jun;123(6):586-592. doi: 10.1111/mmi.15365. Epub 2025 Apr 9. Mol Microbiol. 2025. PMID: 40205788 Free PMC article.
References
-
- Allam A., Kalnis P., and Solovyev V. (2015) ‘Karect: Accurate Correction of Substitution, Insertion and Deletion Errors for Next-generation Sequencing Data’, Bioinformatics, 31: 3421–8. - PubMed
-
- Baker K. E., and Parker R. (2004) ‘Nonsense-mediated mRNA Decay: Terminating Erroneous Gene Expression’, Current Opinion in Cell Biology, 16: 293–9. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

